1RS9
 
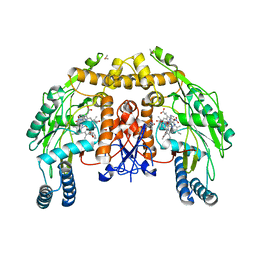 | | Bovine endothelial NOS heme domain with D-phenylalanine-D-nitroarginine amide bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, CACODYLATE ION, ... | | Authors: | Flinspach, M, Li, H, Jamal, J, Yang, W, Huang, H, Silverman, R.B, Poulos, T.L. | | Deposit date: | 2003-12-09 | | Release date: | 2004-06-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structures of the neuronal and endothelial nitric oxide synthase heme domain with D-nitroarginine-containing dipeptide inhibitors bound.
Biochemistry, 43, 2004
|
|
1RS7
 
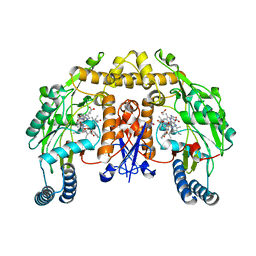 | | Rat neuronal NOS heme domain with D-phenylalanine-D-nitroarginine amide bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, D-MANNITOL, ... | | Authors: | Flinspach, M, Li, H, Jamal, J, Yang, W, Huang, H, Silverman, R.B, Poulos, T.L. | | Deposit date: | 2003-12-09 | | Release date: | 2004-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of the Neuronal and Endothelial Nitric Oxide Synthase Heme Domain with d-Nitroarginine-Containing Dipeptide Inhibitors Bound.
Biochemistry, 43, 2004
|
|
6M11
 
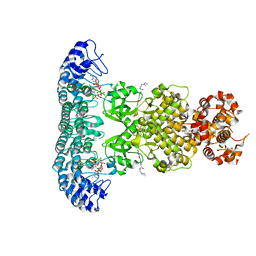 | | Crystal structure of Rnase L in complex with Sunitinib | | Descriptor: | 5'-O-MONOPHOSPHORYLADENYLYL(2'->5')ADENYLYL(2'->5')ADENOSINE, N-[2-(diethylamino)ethyl]-5-[(Z)-(5-fluoro-2-oxo-1,2-dihydro-3H-indol-3-ylidene)methyl]-2,4-dimethyl-1H-pyrrole-3-carbo xamide, PHOSPHATE ION, ... | | Authors: | Tang, J, Huang, H. | | Deposit date: | 2020-02-24 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Sunitinib inhibits RNase L by destabilizing its active dimer conformation.
Biochem.J., 477, 2020
|
|
6M12
 
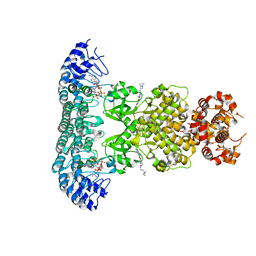 | | Crystal Structure of Rnase L in complex with SU11652 | | Descriptor: | 5-[(E)-(5-CHLORO-2-OXO-1,2-DIHYDRO-3H-INDOL-3-YLIDENE)METHYL]-N-[2-(DIETHYLAMINO)ETHYL]-2,4-DIMETHYL-1H-PYRROLE-3-CARBOXAMIDE, PHOSPHATE ION, Ribonuclease L, ... | | Authors: | Tang, J, Huang, H. | | Deposit date: | 2020-02-24 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Sunitinib inhibits RNase L by destabilizing its active dimer conformation.
Biochem.J., 477, 2020
|
|
6M13
 
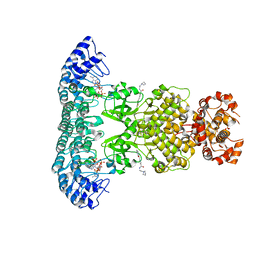 | | Crystal structure of Rnase L in complex with Toceranib | | Descriptor: | 5-[(Z)-(5-fluoranyl-2-oxidanylidene-1H-indol-3-ylidene)methyl]-2,4-dimethyl-N-(2-pyrrolidin-1-ylethyl)-1H-pyrrole-3-carboxamide, PHOSPHATE ION, Ribonuclease L, ... | | Authors: | Tang, J, Huang, H. | | Deposit date: | 2020-02-24 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Sunitinib inhibits RNase L by destabilizing its active dimer conformation.
Biochem.J., 477, 2020
|
|
6U8D
 
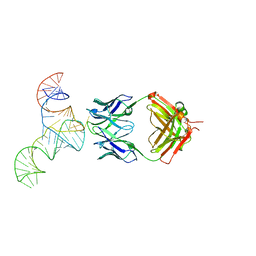 | | Crystal structure of hepatitis C virus IRES junction IIIabc in complex with Fab HCV2 | | Descriptor: | Heavy chain of Fab HCV2, JIIIabc RNA (68-MER), Light chain of Fab HCV2 | | Authors: | Koirala, D, Lewicka, A, Koldobskaya, Y, Huang, H, Piccirilli, J.A. | | Deposit date: | 2019-09-04 | | Release date: | 2019-12-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.807 Å) | | Cite: | Synthetic Antibody Binding to a Preorganized RNA Domain of Hepatitis C Virus Internal Ribosome Entry Site Inhibits Translation.
Acs Chem.Biol., 15, 2020
|
|
6U8K
 
 | | Crystal structure of hepatitis C virus IRES junction IIIabc in complex with Fab HCV3 | | Descriptor: | Heavy chain of Fab HCV3, JIIIabc RNA (68-MER), Light chain of Fab HCV3 | | Authors: | Koirala, D, Lewicka, A, Koldobskaya, Y, Huang, H, Piccirilli, J.A. | | Deposit date: | 2019-09-05 | | Release date: | 2019-12-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Synthetic Antibody Binding to a Preorganized RNA Domain of Hepatitis C Virus Internal Ribosome Entry Site Inhibits Translation.
Acs Chem.Biol., 15, 2020
|
|
3MAZ
 
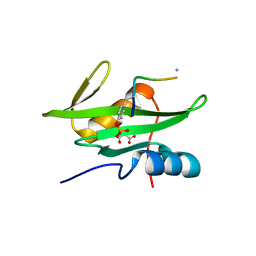 | | Crystal Structure of the Human BRDG1/STAP-1 SH2 Domain in Complex with the NTAL pTyr136 Peptide | | Descriptor: | CheD family protein, MALONATE ION, Signal-transducing adaptor protein 1 | | Authors: | Kaneko, T, Huang, H, Zhao, B, Li, L, Liu, H, Voss, C.K, Wu, C, Schiller, M.R, Li, S.S. | | Deposit date: | 2010-03-24 | | Release date: | 2010-05-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Loops govern SH2 domain specificity by controlling access to binding pockets.
Sci.Signal., 3, 2010
|
|
7YW1
 
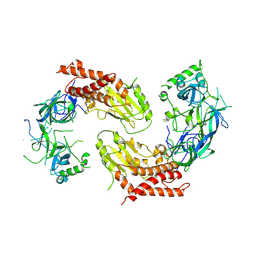 | | crystal structure of UBE2O | | Descriptor: | (E3-independent) E2 ubiquitin-conjugating enzyme UBE2O | | Authors: | Fu, Z, Zhu, W, Huang, H. | | Deposit date: | 2022-08-20 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | crystal structure of UBE2O
To Be Published
|
|
4R4P
 
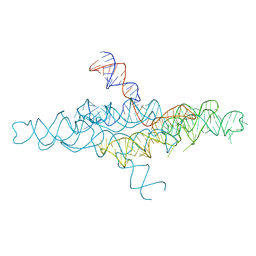 | | Crystal Structure of the VS ribozyme-A756G mutant | | Descriptor: | MAGNESIUM ION, VS ribozyme RNA | | Authors: | Piccirilli, J.A, Suslov, N.B, Dasgupta, S, Huang, H, Lilley, D.M.J, Rice, P.A. | | Deposit date: | 2014-08-19 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Crystal structure of the Varkud satellite ribozyme.
Nat.Chem.Biol., 11, 2015
|
|
4R4V
 
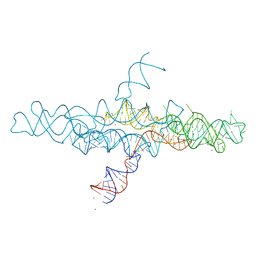 | | Crystal structure of the VS ribozyme - G638A mutant | | Descriptor: | MAGNESIUM ION, POTASSIUM ION, VS ribozyme RNA | | Authors: | Piccirilli, J.A, Suslov, N.B, Dasgupta, S, Huang, H, Lilley, D.M.J, Rice, P.A. | | Deposit date: | 2014-08-19 | | Release date: | 2015-09-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Crystal structure of the Varkud satellite ribozyme.
Nat.Chem.Biol., 11, 2015
|
|
3GYP
 
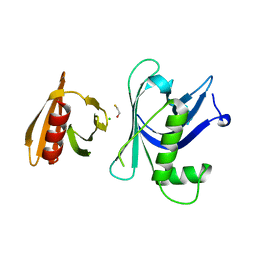 | | Rtt106p | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Histone chaperone RTT106 | | Authors: | Liu, Y, Huang, H, Shi, Y, Teng, M. | | Deposit date: | 2009-04-04 | | Release date: | 2009-12-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.406 Å) | | Cite: | Structural analysis of Rtt106p reveals a DNA-binding role required for heterochromatin silencing
J.Biol.Chem., 285, 2010
|
|
4YLF
 
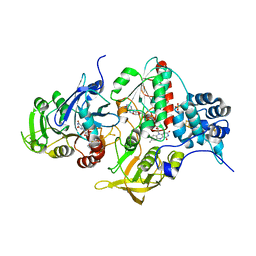 | | Insights into flavin-based electron bifurcation via the NADH-dependent reduced ferredoxin-NADP oxidoreductase structure | | Descriptor: | Dihydroorotate dehydrogenase B (NAD(+)), electron transfer subunit homolog, Dihydropyrimidine dehydrogenase subunit A, ... | | Authors: | Ermler, U, Thauer, R.K, Demmer, J.K, Huang, H, Wang, S, Demmer, U. | | Deposit date: | 2015-03-05 | | Release date: | 2015-07-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Insights into Flavin-based Electron Bifurcation via the NADH-dependent Reduced Ferredoxin:NADP Oxidoreductase Structure.
J.Biol.Chem., 290, 2015
|
|
5IJC
 
 | | The crystal structure of mouse TLR4/MD-2/neoseptin-3 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lymphocyte antigen 96, ... | | Authors: | Wang, Y, Su, L, Morin, M.D, Jones, B.T, Whitby, L.R, Surakattula, M, Huang, H, Shi, H, Choi, J.H, Wang, K, Moresco, E.M, Berger, M, Zhan, X, Zhang, H, Boger, D.L, Beutler, B. | | Deposit date: | 2016-03-01 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | TLR4/MD-2 activation by a synthetic agonist with no similarity to LPS.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5T57
 
 | | Crystal Structure of a Semialdehyde dehydrogenase NAD-binding Protein from Cupriavidus necator in Complex with Calcium and NAD | | Descriptor: | CALCIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Semialdehyde dehydrogenase NAD-binding protein, ... | | Authors: | Cook, W.J, Fedorov, A.A, Fedorov, E.V, Huang, H, Bonanno, J.B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2016-08-30 | | Release date: | 2016-09-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of a Semialdehyde dehydrogenase NAD-binding Protein from Cupriavidus necator in Complex with Calcium and NAD
To be published
|
|
3GYO
 
 | | Se-Met Rtt106p | | Descriptor: | Histone chaperone RTT106 | | Authors: | Liu, Y, Huang, H, Shi, Y, Teng, M. | | Deposit date: | 2009-04-04 | | Release date: | 2009-12-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural analysis of Rtt106p reveals a DNA-binding role required for heterochromatin silencing
J.Biol.Chem., 285, 2010
|
|
4YRY
 
 | | Insights into flavin-based electron bifurcation via the NADH-dependent reduced ferredoxin-NADP oxidoreductase structure | | Descriptor: | Dihydroorotate dehydrogenase B (NAD(+)), electron transfer subunit homolog, Dihydropyrimidine dehydrogenase subunit A, ... | | Authors: | Ermler, U, Thauer, R.K, Demmer, J.K, Huang, H, Wang, S, Demmer, U. | | Deposit date: | 2015-03-16 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Insights into Flavin-based Electron Bifurcation via the NADH-dependent Reduced Ferredoxin:NADP Oxidoreductase Structure.
J.Biol.Chem., 290, 2015
|
|
5IJB
 
 | | The ligand-free structure of the mouse TLR4/MD-2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lymphocyte antigen 96, ... | | Authors: | Wang, Y, Su, L, Morin, M.D, Jones, B.T, Whitby, L.R, Surakattula, M, Huang, H, Shi, H, Choi, J.H, Wang, K, Moresco, E.M, Berger, M, Zhan, X, Zhang, H, Boger, D.L, Beutler, B. | | Deposit date: | 2016-03-01 | | Release date: | 2016-04-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | TLR4/MD-2 activation by a synthetic agonist with no similarity to LPS.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5IJD
 
 | | The crystal structure of mouse TLR4/MD-2/lipid A complex | | Descriptor: | (R)-((2R,3S,4R,5R,6R)-3-HYDROXY-2-(HYDROXYMETHYL)-5-((R)-3-HYDROXYTETRADECANAMIDO)-6-(PHOSPHONOOXY)TETRAHYDRO-2H-PYRAN-4-YL) 3-HYDROXYTETRADECANOATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, Y, Su, L, Morin, M.D, Jones, B.T, Whitby, L.R, Surakattula, M, Huang, H, Shi, H, Choi, J.H, Wang, K, Moresco, E.M, Berger, M, Zhan, X, Zhang, H, Boger, D.L, Beutler, B. | | Deposit date: | 2016-03-01 | | Release date: | 2016-04-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | TLR4/MD-2 activation by a synthetic agonist with no similarity to LPS.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2AX5
 
 | | Solution Structure of Urm1 from Saccharomyces Cerevisiae | | Descriptor: | Hypothetical 11.0 kDa protein in FAA3-MAS3 intergenic region | | Authors: | Xu, J, Huang, H, Zhang, J, Wu, J, Shi, Y. | | Deposit date: | 2005-09-03 | | Release date: | 2006-06-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Urm1 and its implications for the origin of protein modifiers.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
5TNV
 
 | | Crystal Structure of a Xylose isomerase-like TIM barrel Protein from Mycobacterium smegmatis in Complex with Magnesium | | Descriptor: | AP endonuclease, family protein 2, MAGNESIUM ION | | Authors: | Cook, W.J, Fedorov, A.A, Fedorov, E.V, Huang, H, Bonanno, J.B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2016-10-14 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Crystal Structure of a Xylose isomerase-like TIM barrel Protein from Mycobacterium smegmatis in Complex with Magnesium
To Be Published
|
|
3OF7
 
 | | The Crystal Structure of Prp20p from Saccharomyces cerevisiae and Its Binding Properties to Gsp1p and Histones | | Descriptor: | Regulator of chromosome condensation | | Authors: | Wu, F, Liu, Y, Zhu, Z, Huang, H, Ding, B, Wu, J, Shi, Y. | | Deposit date: | 2010-08-14 | | Release date: | 2011-03-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The 1.9A crystal structure of Prp20p from Saccharomyces cerevisiae and its binding properties to Gsp1p and histones.
J.Struct.Biol., 174, 2011
|
|
4F59
 
 | | Triple mutant Src SH2 domain | | Descriptor: | Proto-oncogene tyrosine-protein kinase Src | | Authors: | Kaneko, T, Huang, H, Cao, X, Li, C, Voss, C, Sidhu, S.S, Li, S.S. | | Deposit date: | 2012-05-12 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Superbinder SH2 Domains Act as Antagonists of Cell Signaling.
Sci.Signal., 5, 2012
|
|
4F5A
 
 | | Triple mutant Src SH2 domain bound to phosphate ion | | Descriptor: | PHOSPHATE ION, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Kaneko, T, Huang, H, Cao, X, Li, C, Voss, C, Sidhu, S.S, Li, S.S. | | Deposit date: | 2012-05-12 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Superbinder SH2 Domains Act as Antagonists of Cell Signaling.
Sci.Signal., 5, 2012
|
|
5UHW
 
 | | Crystal Structure of an Oxidoreductase from Agrobacterium radiobacter in Complex with NAD+ and Magnesium | | Descriptor: | MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Oxidoreductase protein | | Authors: | Cook, W.J, Fedorov, A.A, Fedorov, E.V, Huang, H, Bonanno, J.B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2017-01-12 | | Release date: | 2017-01-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystal Structure of an Oxidoreductase from Agrobacterium radiobacter in Complex with NAD+ and Magnesium
To be published
|
|
