4Q35
 
 | | Structure of a membrane protein | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, LAURYL DIMETHYLAMINE-N-OXIDE, LPS-assembly lipoprotein LptE, ... | | Authors: | Huang, Y, Qiao, S, Luo, Q, Zhao, Y. | | Deposit date: | 2014-04-11 | | Release date: | 2014-06-25 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | Structural basis for lipopolysaccharide insertion in the bacterial outer membrane.
Nature, 511, 2014
|
|
4Q79
 
 | | Structure of a HG-derivative CsgG | | Descriptor: | CsgG, MERCURY (II) ION | | Authors: | Huang, Y, Zhang, C.X, Cao, B, Zhao, Y, Kou, Y, Ni, D. | | Deposit date: | 2014-04-24 | | Release date: | 2014-12-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the nonameric bacterial amyloid secretion channel
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
8SRK
 
 | | Cryo-EM structure of TRPM2 chanzyme (without NUDT9-H domain) in the presence of Ca and ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, CALCIUM ION, CHOLESTEROL, ... | | Authors: | Huang, Y, Kumar, S, Lu, W, Du, J. | | Deposit date: | 2023-05-05 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.14 Å) | | Cite: | Coupling enzymatic activity and gating in an ancient TRPM chanzyme and its molecular evolution
To Be Published
|
|
8SRE
 
 | | Cryo-EM structure of TRPM2 chanzyme in the presence of Magnesium and ADP-ribose, closed state | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, CHOLESTEROL, MAGNESIUM ION, ... | | Authors: | Huang, Y, Kumar, S, Lu, W, Du, J. | | Deposit date: | 2023-05-05 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Coupling enzymatic activity and gating in an ancient TRPM chanzyme and its molecular evolution
To Be Published
|
|
8SRI
 
 | | Cryo-EM structure of TRPM2 chanzyme (E1114A) in the presence of Magnesium and ADP-ribose, closed state | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, CHOLESTEROL, MAGNESIUM ION, ... | | Authors: | Huang, Y, Kumar, S, Lu, W, Du, J. | | Deposit date: | 2023-05-05 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Coupling enzymatic activity and gating in an ancient TRPM chanzyme and its molecular evolution
To Be Published
|
|
8SRH
 
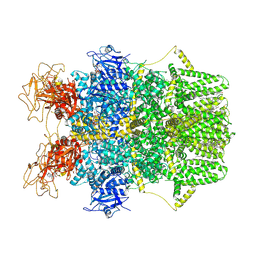 | | Cryo-EM structure of TRPM2 chanzyme (E1114A) in the presence of Magnesium and ADP-ribose, open state | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, CHOLESTEROL, MAGNESIUM ION, ... | | Authors: | Huang, Y, Kumar, S, Lu, W, Du, J. | | Deposit date: | 2023-05-05 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Coupling enzymatic activity and gating in an ancient TRPM chanzyme and its molecular evolution
To Be Published
|
|
8SRG
 
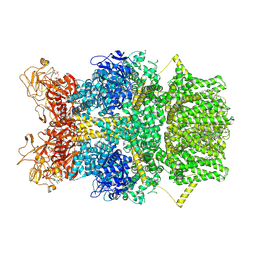 | | Cryo-EM structure of TRPM2 chanzyme in the presence of Magnesium, Adenosine monophosphate, and Ribose-5-phosphate | | Descriptor: | 5-O-phosphono-beta-D-ribofuranose, ADENOSINE MONOPHOSPHATE, CHOLESTEROL, ... | | Authors: | Huang, Y, Kumar, S, Lu, W, Du, J. | | Deposit date: | 2023-05-05 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Coupling enzymatic activity and gating in an ancient TRPM chanzyme and its molecular evolution
To Be Published
|
|
8SRF
 
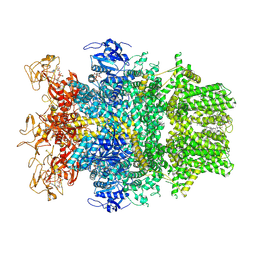 | | Cryo-EM structure of TRPM2 chanzyme in the presence of Magnesium, ADP-ribose, Adenosine monophosphate, and Ribose-5-phosphate, closed state | | Descriptor: | 5-O-phosphono-beta-D-ribofuranose, ADENOSINE MONOPHOSPHATE, ADENOSINE-5-DIPHOSPHORIBOSE, ... | | Authors: | Huang, Y, Kumar, S, Lu, W, Du, J. | | Deposit date: | 2023-05-05 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Coupling enzymatic activity and gating in an ancient TRPM chanzyme and its molecular evolution
To Be Published
|
|
8SRD
 
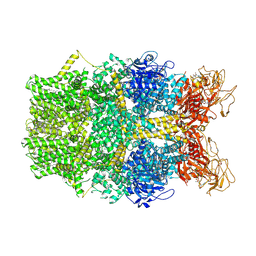 | | Cryo-EM structure of TRPM2 chanzyme in the presence of Magnesium and ADP-ribose, open state | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, CHOLESTEROL, MAGNESIUM ION, ... | | Authors: | Huang, Y, Kumar, S, Lu, W, Du, J. | | Deposit date: | 2023-05-05 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Coupling enzymatic activity and gating in an ancient TRPM chanzyme and its molecular evolution
To Be Published
|
|
7CFG
 
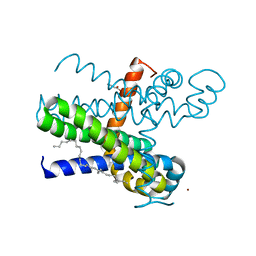 | | Structure of the transmembrane domain of the bacterial CNNM/CorC family Mg2+ transporter in complex with Mg2+ | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Hemolysin, MAGNESIUM ION, ... | | Authors: | Huang, Y, Jin, F, Hattori, M. | | Deposit date: | 2020-06-25 | | Release date: | 2021-02-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for the Mg 2+ recognition and regulation of the CorC Mg 2+ transporter.
Sci Adv, 7, 2021
|
|
7CFF
 
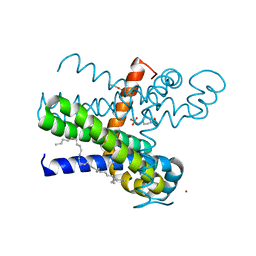 | | Structure of the thermostabilized transmembrane domain of the bacterial CNNM/CorC family Mg2+ transporter in complex with Mg2+ | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Hemolysin, ... | | Authors: | Huang, Y, Jin, F, Hattori, M. | | Deposit date: | 2020-06-25 | | Release date: | 2021-02-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the Mg 2+ recognition and regulation of the CorC Mg 2+ transporter.
Sci Adv, 7, 2021
|
|
7CFI
 
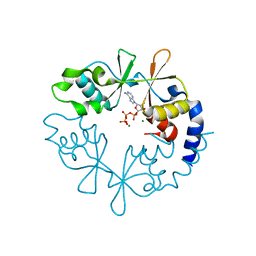 | |
8YLC
 
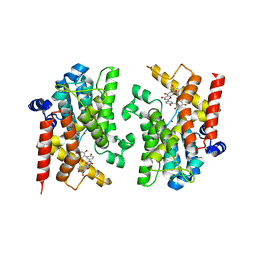 | | The crystal structure of PDE4D with Amentoflavone | | Descriptor: | 3',5'-cyclic-AMP phosphodiesterase 4D, 8-[5-[5,7-bis(oxidanyl)-4-oxidanylidene-chromen-2-yl]-2-oxidanyl-phenyl]-2-(4-hydroxyphenyl)-5,7-bis(oxidanyl)chromen-4-one, MAGNESIUM ION, ... | | Authors: | Huang, Y.-Y, Luo, H.-B. | | Deposit date: | 2024-03-06 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.30003715 Å) | | Cite: | Discovery of amentoflavone as a natural PDE4 inhibitor with anti-fibrotic effects
Chin.Chem.Lett., 2024
|
|
6TQN
 
 | | rrn anti-termination complex without S4 | | Descriptor: | 30S ribosomal protein S10, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Huang, Y.H, Wahl, M.C, Loll, B, Hilal, T, Said, N. | | Deposit date: | 2019-12-17 | | Release date: | 2020-08-05 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure-Based Mechanisms of a Molecular RNA Polymerase/Chaperone Machine Required for Ribosome Biosynthesis.
Mol.Cell, 79, 2020
|
|
6TQO
 
 | | rrn anti-termination complex | | Descriptor: | 30S ribosomal protein S10, 30S ribosomal protein S4, DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Huang, Y.H, Wahl, M.C, Loll, B, Hilal, T, Said, N. | | Deposit date: | 2019-12-17 | | Release date: | 2020-08-05 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure-Based Mechanisms of a Molecular RNA Polymerase/Chaperone Machine Required for Ribosome Biosynthesis.
Mol.Cell, 79, 2020
|
|
4QXW
 
 | | Crystal structure of the human CEACAM1 membrane distal amino terminal (N)-domain | | Descriptor: | Carcinoembryonic antigen-related cell adhesion molecule 1, MALONIC ACID, octyl beta-D-glucopyranoside | | Authors: | Huang, Y.H, Gandhi, A.K, Russell, A, Kondo, Y, Chen, Q, Petsko, G.A, Blumberg, R.S. | | Deposit date: | 2014-07-22 | | Release date: | 2014-11-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | CEACAM1 regulates TIM-3-mediated tolerance and exhaustion.
Nature, 517, 2015
|
|
7RMR
 
 | | Crystal structure of [I11L]cycloviolacin O2 | | Descriptor: | D-[I11L]cycloviolacin O2, THIOCYANATE ION, [I11L]cycloviolacin O2 | | Authors: | Huang, Y.H, Du, Q, Craik, D.J. | | Deposit date: | 2021-07-28 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Enabling Efficient Folding and High-Resolution Crystallographic Analysis of Bracelet Cyclotides.
Molecules, 26, 2021
|
|
7RMQ
 
 | | Crystal structure of cycloviolacin O2 | | Descriptor: | Cycloviolacin O2, D-[I11L]cycloviolacin O2, FORMIC ACID | | Authors: | Huang, Y.H, Du, Q. | | Deposit date: | 2021-07-28 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Enabling Efficient Folding and High-Resolution Crystallographic Analysis of Bracelet Cyclotides.
Molecules, 26, 2021
|
|
7RMS
 
 | | Crystal structure of [I11G]cycloviolacin O2 | | Descriptor: | D-[I11L]cycloviolacin O2, THIOCYANATE ION, [I11L]cycloviolacin O2 | | Authors: | Huang, Y.H, Du, Q, Craik, D.J. | | Deposit date: | 2021-07-28 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Enabling Efficient Folding and High-Resolution Crystallographic Analysis of Bracelet Cyclotides.
Molecules, 26, 2021
|
|
8SRA
 
 | | Cryo-EM structure of TRPM2 chanzyme in the presence of Calcium | | Descriptor: | CALCIUM ION, CHOLESTEROL, TRPM2 chanzyme | | Authors: | Huang, Y, Kumar, S, Lu, W, Du, J. | | Deposit date: | 2023-05-05 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Coupling enzymatic activity and gating in an ancient TRPM chanzyme and its molecular evolution.
Nat.Struct.Mol.Biol., 2024
|
|
8SR9
 
 | | Cryo-EM structure of TRPM2 chanzyme in the presence of Magnesium | | Descriptor: | CHOLESTEROL, MAGNESIUM ION, TRPM2 chanzyme | | Authors: | Huang, Y, Kumar, S, Lu, W, Du, J. | | Deposit date: | 2023-05-05 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Coupling enzymatic activity and gating in an ancient TRPM chanzyme and its molecular evolution.
Nat.Struct.Mol.Biol., 2024
|
|
8SR8
 
 | |
8SRB
 
 | | Cryo-EM structure of TRPM2 chanzyme in the presence of EDTA and ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, CHOLESTEROL, TRPM2 chanzyme | | Authors: | Huang, Y, Kumar, S, Lu, W, Du, J. | | Deposit date: | 2023-05-05 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Coupling enzymatic activity and gating in an ancient TRPM chanzyme and its molecular evolution.
Nat.Struct.Mol.Biol., 2024
|
|
7CFH
 
 | |
1KKG
 
 | | NMR Structure of Ribosome-Binding Factor A (RbfA) | | Descriptor: | ribosome-binding factor A | | Authors: | Huang, Y.J, Swapna, G.V.T, Rajan, P.K, Ke, H, Xia, B, Shukla, K, Inouye, M, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2001-12-07 | | Release date: | 2003-03-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of Ribosome-binding Factor A (RbfA), A Cold-shock
Adaptation Protein from Escherichia coli
J.Mol.Biol., 327, 2003
|
|
