8HK2
 
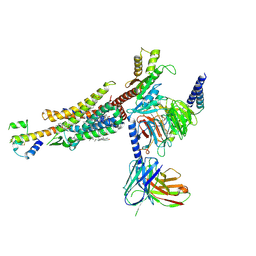 | | C3aR-Gi-C3a protein complex | | Descriptor: | C3a anaphylatoxin, C3a anaphylatoxin chemotactic receptor, CHOLESTEROL, ... | | Authors: | Wang, Y, Liu, W, Xu, Y, Zhuang, Y, Xu, H.E. | | Deposit date: | 2022-11-24 | | Release date: | 2023-05-10 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Revealing the signaling of complement receptors C3aR and C5aR1 by anaphylatoxins.
Nat.Chem.Biol., 19, 2023
|
|
7TIW
 
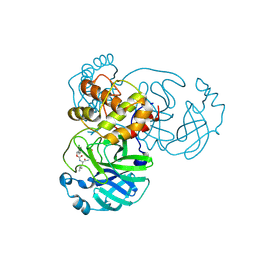 | | Crystal structure of SARS-CoV-2 3CL in complex with inhibitor EB54 | | Descriptor: | (1S,2S)-2-[(N-{[(2-chlorophenyl)methoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5, PHOSPHATE ION | | Authors: | Forouhar, F, Liu, H, Iketani, S, Zack, A, Khanizeman, N, Bednarova, E, Fowler, B, Hong, S.J, Mohri, H, Nair, M.S, Huang, Y, Tay, N.E.S, Lee, S, Karan, C, Resnick, S.J, Quinn, C, Li, W, Shion, H, Jurtschenko, C, Lauber, M.A, McDonald, T, Stokes, M.E, Hurst, B, Rovis, T, Chavez, A, Ho, D.D, Stockwell, B.R. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Development of optimized drug-like small molecule inhibitors of the SARS-CoV-2 3CL protease for treatment of COVID-19.
Nat Commun, 13, 2022
|
|
7TIA
 
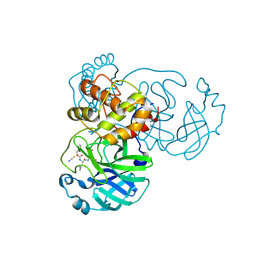 | | Crystal structure of SARS-CoV-2 3CL in complex with inhibitor NK01-14 | | Descriptor: | 3C-like proteinase nsp5, THIOCYANATE ION, benzyl [(2S)-3-cyclopropyl-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-1-oxopropan-2-yl]carbamate | | Authors: | Forouhar, F, Liu, H, Iketani, S, Zack, A, Khanizeman, N, Bednarova, E, Fowler, B, Hong, S.J, Mohri, H, Nair, M.S, Huang, Y, Tay, N.E.S, Lee, S, Karan, C, Resnick, S.J, Quinn, C, Li, W, Shion, H, Jurtschenko, C, Lauber, M.A, McDonald, T, Stokes, M.E, Hurst, B, Rovis, T, Chavez, A, Ho, D.D, Stockwell, B.R. | | Deposit date: | 2022-01-13 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Development of optimized drug-like small molecule inhibitors of the SARS-CoV-2 3CL protease for treatment of COVID-19.
Nat Commun, 13, 2022
|
|
7TIX
 
 | | Crystal structure of SARS-CoV-2 3CL in complex with inhibitor EB56 | | Descriptor: | 3C-like proteinase nsp5, MAGNESIUM ION, N~2~-{[(naphthalen-2-yl)methoxy]carbonyl}-N-{(2S)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Forouhar, F, Liu, H, Iketani, S, Zack, A, Khanizeman, N, Bednarova, E, Fowler, B, Hong, S.J, Mohri, H, Nair, M.S, Huang, Y, Tay, N.E.S, Lee, S, Karan, C, Resnick, S.J, Quinn, C, Li, W, Shion, H, Jurtschenko, C, Lauber, M.A, McDonald, T, Stokes, M.E, Hurst, B, Rovis, T, Chavez, A, Ho, D.D, Stockwell, B.R. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Development of optimized drug-like small molecule inhibitors of the SARS-CoV-2 3CL protease for treatment of COVID-19.
Nat Commun, 13, 2022
|
|
7TJ0
 
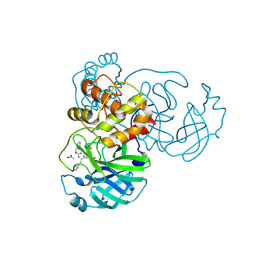 | | Crystal structure of SARS-CoV-2 3CL in complex with inhibitor SL-4-241 | | Descriptor: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-3-cyclohexyl-L-alanyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5, ACETATE ION | | Authors: | Forouhar, F, Liu, H, Iketani, S, Zack, A, Khanizeman, N, Bednarova, E, Fowler, B, Hong, S.J, Mohri, H, Nair, M.S, Huang, Y, Tay, N.E.S, Lee, S, Karan, C, Resnick, S.J, Quinn, C, Li, W, Shion, H, Jurtschenko, C, Lauber, M.A, McDonald, T, Stokes, M, Hurst, B, Rovis, T, Chavez, A, Ho, D.D, Stockwell, B.R. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Development of optimized drug-like small molecule inhibitors of the SARS-CoV-2 3CL protease for treatment of COVID-19.
Nat Commun, 13, 2022
|
|
7RN3
 
 | | hyen D solution structure | | Descriptor: | Cyclotide hyen-D | | Authors: | Du, Q, Huang, Y.H, Craik, D.J, Wang, C.K. | | Deposit date: | 2021-07-29 | | Release date: | 2022-03-02 | | Last modified: | 2022-12-07 | | Method: | SOLUTION NMR | | Cite: | Mutagenesis of bracelet cyclotide hyen D reveals functionally and structurally critical residues for membrane binding and cytotoxicity.
J.Biol.Chem., 298, 2022
|
|
1D4F
 
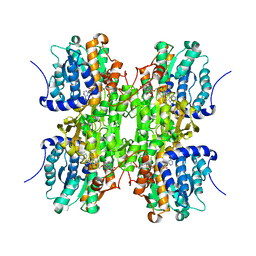 | | CRYSTAL STRUCTURE OF RECOMBINANT RAT-LIVER D244E MUTANT S-ADENOSYLHOMOCYSTEINE HYDROLASE | | Descriptor: | ADENOSINE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, S-ADENOSYLHOMOCYSTEINE HYDROLASE | | Authors: | Komoto, J, Huang, Y, Takusagawa, F, Gomi, T, Ogawa, H, Takata, Y, Fujioka, M. | | Deposit date: | 2000-06-22 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Effects of site-directed mutagenesis on structure and function of recombinant rat liver S-adenosylhomocysteine hydrolase. Crystal structure of D244E mutant enzyme.
J.Biol.Chem., 275, 2000
|
|
4U68
 
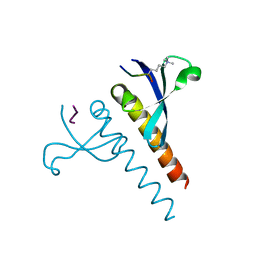 | |
7D47
 
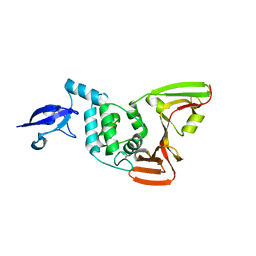 | | Crystal structure of SARS-CoV-2 Papain-like protease C111S | | Descriptor: | CALCIUM ION, Non-structural protein 3, ZINC ION | | Authors: | Wu, K.-P, Chen, S.-K, Lu, Y.-C, Huang, Y.-C.J, Lee, M.-H. | | Deposit date: | 2020-09-22 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of SARS-CoV-2 Papain-like protease
To Be Published
|
|
1Z5F
 
 | | Solution Structure of the Cytotoxic RC-RNase 3 with a Pyroglutamate Residue at the N-terminus | | Descriptor: | RC-RNase 3 | | Authors: | Lou, Y.C, Huang, Y.C, Pan, Y.R, Chen, C, Liao, Y.D. | | Deposit date: | 2005-03-18 | | Release date: | 2006-02-28 | | Last modified: | 2019-12-25 | | Method: | SOLUTION NMR | | Cite: | Roles of N-terminal pyroglutamate in maintaining structural integrity and pKa values of catalytic histidine residues in bullfrog ribonuclease 3
J.Mol.Biol., 355, 2006
|
|
4PL6
 
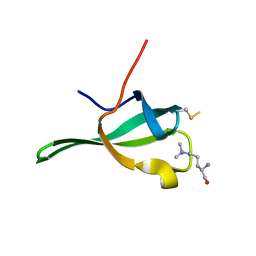 | |
6LYS
 
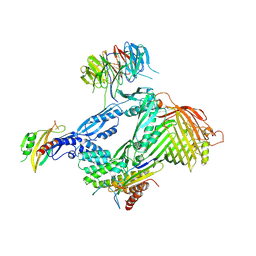 | | Structure of the BAM complex | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Xiao, L, Huang, Y. | | Deposit date: | 2020-02-15 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structures of the beta-barrel assembly machine recognizing outer membrane protein substrates.
Faseb J., 35, 2021
|
|
3NTI
 
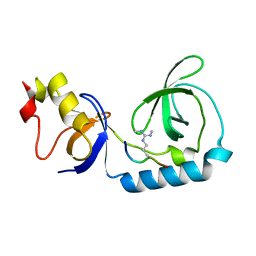 | | Crystal structure of Tudor and Aubergine [R15(me2s)] complex | | Descriptor: | Maternal protein tudor, peptide from Aubergine | | Authors: | Liu, H.P, Huang, Y, Li, Z.Z, Gong, W.M, Xu, R.M. | | Deposit date: | 2010-07-05 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for methylarginine-dependent recognition of Aubergine by Tudor
Genes Dev., 24, 2010
|
|
3NTK
 
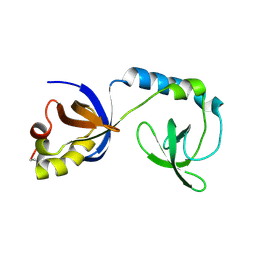 | | Crystal structure of Tudor | | Descriptor: | Maternal protein tudor | | Authors: | Liu, H.P, Huang, Y, Li, Z.Z, Gong, W.M, Xu, R.M. | | Deposit date: | 2010-07-05 | | Release date: | 2010-09-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for methylarginine-dependent recognition of Aubergine by Tudor
Genes Dev., 24, 2010
|
|
2V0X
 
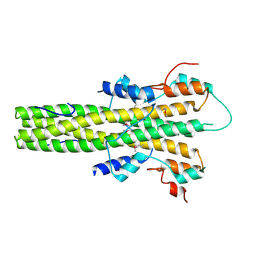 | | The dimerization domain of LAP2alpha | | Descriptor: | LAMINA-ASSOCIATED POLYPEPTIDE 2 ISOFORMS ALPHA/ZETA | | Authors: | Bradley, C.M, Jones, S, Huang, Y, Suzuki, Y, Kvaratskhelia, M, Hickman, A.B, Craigie, R, Dyda, F. | | Deposit date: | 2007-05-20 | | Release date: | 2007-06-26 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Dimerization of Lap2Alpha, a Component of the Nuclear Lamina.
Structure, 15, 2007
|
|
1K0U
 
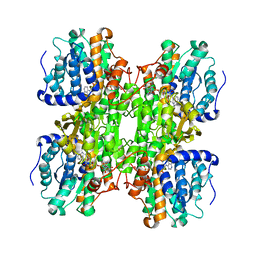 | | Inhibition of S-adenosylhomocysteine Hydrolase by "acyclic sugar" Adenosine Analogue D-eritadenine | | Descriptor: | D-ERITADENINE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, S-ADENOSYL-L-HOMOCYSTEINE HYDROLASE | | Authors: | Takusagawa, F, Huang, Y, Komoto, J, Takata, Y, Gomi, T, Ogawa, H, Fujioka, M, Powell, D. | | Deposit date: | 2001-09-20 | | Release date: | 2001-10-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Inhibition of S-adenosylhomocysteine hydrolase by acyclic sugar adenosine analogue D-eritadenine. Crystal structure of S-adenosylhomocysteine hydrolase complexed with D-eritadenine.
J.Biol.Chem., 277, 2002
|
|
6PUS
 
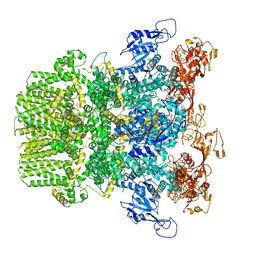 | | Human TRPM2 bound to ADPR and calcium | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, CALCIUM ION, Transient receptor potential cation channel subfamily M member 2 | | Authors: | Du, J, Lu, W, Huang, Y. | | Deposit date: | 2019-07-18 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Ligand recognition and gating mechanism through three ligand-binding sites of human TRPM2 channel.
Elife, 8, 2019
|
|
6PUR
 
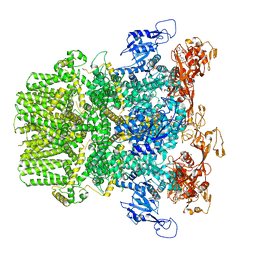 | | Human TRPM2 bound to ADPR | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Transient receptor potential cation channel subfamily M member 2 | | Authors: | Du, J, Lu, W, Huang, Y. | | Deposit date: | 2019-07-18 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Ligand recognition and gating mechanism through three ligand-binding sites of human TRPM2 channel.
Elife, 8, 2019
|
|
6PUU
 
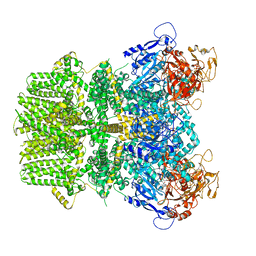 | | Human TRPM2 bound to 8-Br-cADPR and calcium | | Descriptor: | (2R,3R,4S,5R,13R,14S,15R,16R)-24-amino-18-bromo-3,4,14,15-tetrahydroxy-7,9,11,25,26-pentaoxa-17,19,22-triaza-1-azonia-8 ,10-diphosphapentacyclo[18.3.1.1^2,5^.1^13,16^.0^17,21^]hexacosa-1(24),18,20,22-tetraene-8,10-diolate 8,10-dioxide, CALCIUM ION, Transient receptor potential cation channel subfamily M member 2 | | Authors: | Du, J, Lu, W, Huang, Y. | | Deposit date: | 2019-07-18 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Ligand recognition and gating mechanism through three ligand-binding sites of human TRPM2 channel.
Elife, 8, 2019
|
|
8H4R
 
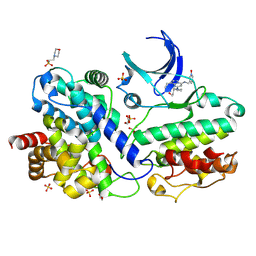 | | The Crystal Structure of CDK3 and CyclinE1 Complex with Dinaciclib from Biortus | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-[({3-ethyl-5-[(2S)-2-(2-hydroxyethyl)piperidin-1-yl]pyrazolo[1,5-a]pyrimidin-7-yl}amino)methyl]-1-hydroxypyridinium, G1/S-specific cyclin-E1, ... | | Authors: | Gui, W, Wang, F, Cheng, W, Gao, J, Huang, Y, Ouyang, Z. | | Deposit date: | 2022-10-11 | | Release date: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The Crystal Structure of CDK3 and CyclinE1 Complex with Dinaciclib from Biortus
To Be Published
|
|
6PUO
 
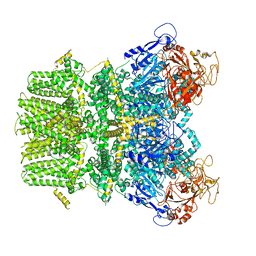 | | Human TRPM2 in the apo state | | Descriptor: | Transient receptor potential cation channel subfamily M member 2 | | Authors: | Du, J, Lu, W, Huang, Y. | | Deposit date: | 2019-07-18 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Ligand recognition and gating mechanism through three ligand-binding sites of human TRPM2 channel.
Elife, 8, 2019
|
|
1NY4
 
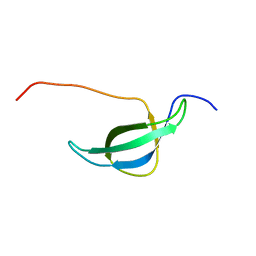 | | Solution structure of the 30S ribosomal protein S28E from Pyrococcus horikoshii. Northeast Structural Genomics Consortium target JR19. | | Descriptor: | 30S ribosomal protein S28E | | Authors: | Aramini, J.M, Cort, J.R, Huang, Y.J, Xiao, R, Acton, T.B, Ho, C.K, Shih, L.-Y, Kennedy, M.A, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-02-11 | | Release date: | 2003-09-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the 30S ribosomal protein S28E from Pyrococcus horikoshii.
Protein Sci., 12, 2003
|
|
1YH5
 
 | | Solution NMR Structure of Protein yggU from Escherichia coli. Northeast Structural Genomics Consortium Target ER14. | | Descriptor: | ORF, HYPOTHETICAL PROTEIN | | Authors: | Aramini, J.M, Xiao, R, Huang, Y.J, Acton, T.B, Wu, M.J, Mills, J.L, Tejero, R.T, Szyperski, T, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-01-06 | | Release date: | 2005-02-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Hypothetical Protein Yggu from E. Coli. Northeast Structural Genomics Consortium Target Er14.
To be Published
|
|
3NTH
 
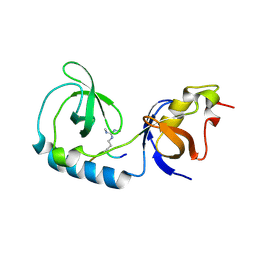 | | Crystal structure of Tudor and Aubergine [R13(me2s)] complex | | Descriptor: | Maternal protein tudor, peptide from Aubergine | | Authors: | Liu, H.P, Huang, Y, Li, Z.Z, Gong, W.M, Xu, R.M. | | Deposit date: | 2010-07-05 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for methylarginine-dependent recognition of Aubergine by Tudor
Genes Dev., 24, 2010
|
|
6L0I
 
 | | Crystal structure of dihydroorotase in complex with malate at pH6.5 from Saccharomyces cerevisiae | | Descriptor: | (2S)-2-hydroxybutanedioic acid, Dihydroorotase, ZINC ION | | Authors: | Guan, H.H, Huang, Y.H, Huang, C.Y, Chen, C.J. | | Deposit date: | 2019-09-26 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the interaction modes of dihydroorotase with the anticancer drugs 5-fluorouracil and 5-aminouracil.
Biochem.Biophys.Res.Commun., 551, 2021
|
|
