4U0I
 
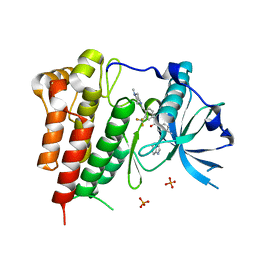 | | Crystal structure of KIT in complex with ponatinib | | Descriptor: | 3-(imidazo[1,2-b]pyridazin-3-ylethynyl)-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzam ide, Mast/stem cell growth factor receptor Kit,Mast/stem cell growth factor receptor Kit, PHOSPHATE ION | | Authors: | Zhou, T, Zhu, X, Dalgarno, D.C. | | Deposit date: | 2014-07-11 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ponatinib Inhibits Polyclonal Drug-Resistant KIT Oncoproteins and Shows Therapeutic Potential in Heavily Pretreated Gastrointestinal Stromal Tumor (GIST) Patients.
Clin.Cancer Res., 20, 2014
|
|
6URM
 
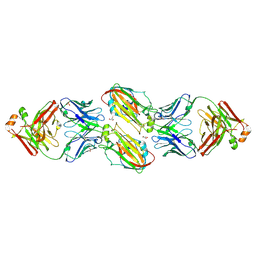 | | Crystal structure of vaccine-elicited receptor-binding site targeting antibody LPAF-a.01 in complex with Hemagglutinin H1 A/California/04/2009 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Zhou, T, Cheung, S.F, Kwong, P.D. | | Deposit date: | 2019-10-23 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Identification and Structure of a Multidonor Class of Head-Directed Influenza-Neutralizing Antibodies Reveal the Mechanism for Its Recurrent Elicitation.
Cell Rep, 32, 2020
|
|
7JWY
 
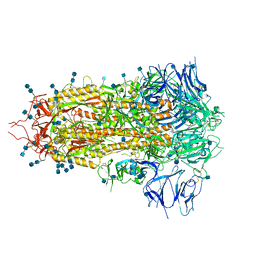 | | Structure of SARS-CoV-2 spike at pH 4.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhou, T, Tsybovsky, Y, Kwong, P.D. | | Deposit date: | 2020-08-26 | | Release date: | 2020-11-25 | | Last modified: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
6NZ7
 
 | |
6BF4
 
 | |
7NIP
 
 | |
7LRT
 
 | |
7LRS
 
 | |
9BIB
 
 | | Rat GluN1-GluN2B NMDA receptor channel in complex with glycine, glutamate, and EU-1622-A, in open-channel conformation, C1 symmetry | | Descriptor: | GLUTAMIC ACID, GLYCINE, Glutamate receptor, ... | | Authors: | Chou, T.-H, Furukawa, H. | | Deposit date: | 2024-04-23 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Molecular mechanism of ligand gating and opening of NMDA receptor.
Nature, 632, 2024
|
|
8T1G
 
 | |
4XAK
 
 | | Crystal structure of potent neutralizing antibody m336 in complex with MERS Co-V RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Heavy chain of neutralizing antibody m336, ... | | Authors: | Zhou, T, Dimtrov, D.S, Ying, T. | | Deposit date: | 2014-12-15 | | Release date: | 2015-08-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Junctional and allele-specific residues are critical for MERS-CoV neutralization by an exceptionally potent germline-like antibody.
Nat Commun, 6, 2015
|
|
4XNZ
 
 | | Crystal structure of broadly and potently neutralizing antibody VRC06B in complex with HIV-1 clade A/E strain 93TH057 gp120 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160,Envelope glycoprotein gp160, ... | | Authors: | Zhou, T, Srivatsan, S, Kwong, P.D. | | Deposit date: | 2015-01-16 | | Release date: | 2015-04-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.389 Å) | | Cite: | Maturation and Diversity of the VRC01-Antibody Lineage over 15 Years of Chronic HIV-1 Infection.
Cell, 161, 2015
|
|
4XMP
 
 | | Crystal structure of broadly and potently neutralizing antibody VRC08 in complex with HIV-1 clade A strain Q842.d12 gp120 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160,Envelope glycoprotein gp160,Envelope glycoprotein gp160, ... | | Authors: | Zhou, T, Srivatsan, S, Kwong, P.D. | | Deposit date: | 2015-01-15 | | Release date: | 2015-04-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7831 Å) | | Cite: | Maturation and Diversity of the VRC01-Antibody Lineage over 15 Years of Chronic HIV-1 Infection.
Cell, 161, 2015
|
|
4XNY
 
 | | Crystal structure of broadly and potently neutralizing antibody VRC08C in complex with HIV-1 clade A strain Q842.d12 gp120 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160,Envelope glycoprotein gp160,Envelope glycoprotein gp160, HEAVY CHAIN OF ANTIBODY VRC08C, ... | | Authors: | Zhou, T, Srivatsan, S, Kwong, P.D. | | Deposit date: | 2015-01-16 | | Release date: | 2015-04-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Maturation and Diversity of the VRC01-Antibody Lineage over 15 Years of Chronic HIV-1 Infection.
Cell, 161, 2015
|
|
7JKS
 
 | | Crystal structure of vaccine-elicited broadly neutralizing VRC01-class antibody 2411a in complex with HIV-1 gp120 core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 gp120 core, The heavy chain of antibody 2411a, ... | | Authors: | Zhou, T, Chen, X, Kwong, P.D, Mascola, J.R. | | Deposit date: | 2020-07-28 | | Release date: | 2021-06-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Vaccination induces maturation in a mouse model of diverse unmutated VRC01-class precursors to HIV-neutralizing antibodies with >50% breadth.
Immunity, 54, 2021
|
|
7JKT
 
 | | Crystal structure of vaccine-elicited broadly neutralizing VRC01-class antibody 2413a in complex with HIV-1 gp120 core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HIV-1 gp120 core from strain d45-01dG5, ... | | Authors: | Zhou, T, Chen, X, Kwong, P.D, Mascola, J.R. | | Deposit date: | 2020-07-28 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Vaccination induces maturation in a mouse model of diverse unmutated VRC01-class precursors to HIV-neutralizing antibodies with >50% breadth.
Immunity, 54, 2021
|
|
7L77
 
 | |
7L79
 
 | |
8TNH
 
 | |
8TNI
 
 | | Cryo-EM structure of HIV-1 Env BG505 DS-SOSIP in complex with broadly neutralizing bi-specific antibody CAP256L-R27 targeting the CD4-binding site and the V2-apex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 BG505 DS-SOSIP gp120, ... | | Authors: | Zhou, T, Morano, N.C, Roark, R.S, Kwong, P.D, Xu, J. | | Deposit date: | 2023-08-01 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Cryo-EM structure of HIV-1 Env BG505 DS-SOSIP in complex with broadly neutralizing bi-specific antibody CAP256L-R27 targeting the CD4-binding site and the V2-apex
To Be Published
|
|
8TNG
 
 | |
7TB4
 
 | | Cryo-EM structure of the spike of SARS-CoV-2 Omicron variant of concern | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Surface glycoprotein | | Authors: | Zhou, T, Tsybovsky, T, Kwong, P.D. | | Deposit date: | 2021-12-21 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Antibodies with potent and broad neutralizing activity against antigenically diverse and highly transmissible SARS-CoV-2 variants.
Biorxiv, 2021
|
|
8FBW
 
 | | Crystal structure of SIV-1 V2 antibody NCI05 in complex with a V2 peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhou, T, Kwong, P.D, Van Wazer, D.J. | | Deposit date: | 2022-11-30 | | Release date: | 2023-01-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Effect of Passive Administration of Monoclonal Antibodies Recognizing Simian Immunodeficiency Virus (SIV) V2 in CH59-Like Coil/Helical or beta-Sheet Conformations on Time of SIV mac251 Acquisition.
J.Virol., 97, 2023
|
|
5F96
 
 | |
7TBF
 
 | | Locally refined region of SARS-CoV-2 spike in complex with antibodies B1-182.1 and A19-61.1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of SARS-CoV-2 antibody A19-61.1, Heavy chain of SARS-CoV-2 antibody B1-182.1, ... | | Authors: | Zhou, T, Tsybovsky, T, Kwong, P.D. | | Deposit date: | 2021-12-21 | | Release date: | 2022-03-30 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for potent antibody neutralization of SARS-CoV-2 variants including B.1.1.529.
Science, 376, 2022
|
|
