8GHJ
 
 | |
8OEE
 
 | |
6O0W
 
 | | Crystal structure of the TIR domain from the grapevine disease resistance protein RUN1 in complex with NADP+ and Bis-Tris | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-2'-5'-DIPHOSPHATE, TIR-NB-LRR type resistance protein RUN1 | | Authors: | Horsefield, S, Burdett, H, Zhang, X, Manik, M.K, Shi, Y, Chen, J, Tiancong, Q, Gilley, J, Lai, J, Gu, W, Rank, M, Casey, L, Ericsson, D.J, Foley, G, Hughes, R.O, Bosanac, T, von Itzstein, M, Rathjen, J.P, Nanson, J.D, Boden, M, Dry, I.B, Williams, S.J, Staskawicz, B.J, Coleman, M.P, Ve, T, Dodds, P.N, Kobe, B. | | Deposit date: | 2019-02-17 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | NAD+cleavage activity by animal and plant TIR domains in cell death pathways.
Science, 365, 2019
|
|
6O0S
 
 | | Crystal structure of the tandem SAM domains from human SARM1 | | Descriptor: | Sterile alpha and TIR motif-containing protein 1 | | Authors: | Horsefield, S, Burdett, H, Zhang, X, Manik, M.K, Shi, Y, Chen, J, Tiancong, Q, Gilley, J, Lai, J, Gu, W, Rank, M, Deerain, N, Casey, L, Ericsson, D.J, Foley, G, Hughes, R.O, Bosanac, T, von Itzstein, M, Rathjen, J.P, Nanson, J.D, Boden, M, Dry, I.B, Williams, S.J, Staskawicz, B.J, Coleman, M.P, Ve, T, Dodds, P.N, Kobe, B. | | Deposit date: | 2019-02-17 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | NAD+cleavage activity by animal and plant TIR domains in cell death pathways.
Science, 365, 2019
|
|
6O0R
 
 | | Crystal structure of the TIR domain from human SARM1 in complex with glycerol | | Descriptor: | GLYCEROL, Sterile alpha and TIR motif-containing protein 1 | | Authors: | Horsefield, S, Burdett, H, Zhang, X, Manik, M.K, Shi, Y, Chen, J, Tiancong, Q, Gilley, J, Lai, J, Gu, W, Rank, M, Deerain, N, Casey, L, Ericsson, D.J, Foley, G, Hughes, R.O, Bosanac, T, von Itzstein, M, Rathjen, J.P, Nanson, J.D, Boden, M, Dry, I.B, Williams, S.J, Staskawicz, B.J, Coleman, M.P, Ve, T, Dodds, P.N, Kobe, B. | | Deposit date: | 2019-02-17 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | NAD+cleavage activity by animal and plant TIR domains in cell death pathways.
Science, 365, 2019
|
|
6O0U
 
 | | Crystal structure of the TIR domain H685A mutant from human SARM1 | | Descriptor: | Sterile alpha and TIR motif-containing protein 1 | | Authors: | Horsefield, S, Burdett, H, Zhang, X, Manik, M.K, Shi, Y, Chen, J, Tiancong, Q, Gilley, J, Lai, J, Gu, W, Rank, M, Casey, L, Ericsson, D.J, Foley, G, Hughes, R.O, Bosanac, T, von Itzstein, M, Rathjen, J.P, Nanson, J.D, Boden, M, Dry, I.B, Williams, S.J, Staskawicz, B.J, Coleman, M.P, Ve, T, Dodds, P.N, Kobe, B. | | Deposit date: | 2019-02-17 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | NAD+cleavage activity by animal and plant TIR domains in cell death pathways.
Science, 365, 2019
|
|
6O0Q
 
 | | Crystal structure of the TIR domain from human SARM1 in complex with ribose | | Descriptor: | CHLORIDE ION, Sterile alpha and TIR motif-containing protein 1, beta-D-ribofuranose | | Authors: | Horsefield, S, Burdett, H, Zhang, X, Manik, M.K, Shi, Y, Chen, J, Tiancong, Q, Gilley, J, Lai, J, Gu, W, Rank, M, Deerain, N, Casey, L, Ericsson, D.J, Foley, G, Hughes, R.O, Bosanac, T, von Itzstein, M, Rathjen, J.P, Nanson, J.D, Boden, M, Dry, I.B, Williams, S.J, Staskawicz, B.J, Coleman, M.P, Ve, T, Dodds, P.N, Kobe, B. | | Deposit date: | 2019-02-17 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | NAD+cleavage activity by animal and plant TIR domains in cell death pathways.
Science, 365, 2019
|
|
6O0T
 
 | | Crystal structure of selenomethionine labelled tandem SAM domains (L446M:L505M:L523M mutant) from human SARM1 | | Descriptor: | Sterile alpha and TIR motif-containing protein 1 | | Authors: | Horsefield, S, Burdett, H, Zhang, X, Manik, M.K, Shi, Y, Chen, J, Tiancong, Q, Gilley, J, Lai, J, Gu, W, Rank, M, Deerain, N, Casey, L, Ericsson, D.J, Foley, G, Hughes, R.O, Bosanac, T, von Itzstein, M, Rathjen, J.P, Nanson, J.D, Boden, M, Dry, I.B, Williams, S.J, Staskawicz, B.J, Coleman, M.P, Ve, T, Dodds, P.N, Kobe, B. | | Deposit date: | 2019-02-17 | | Release date: | 2019-09-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | NAD+cleavage activity by animal and plant TIR domains in cell death pathways.
Science, 365, 2019
|
|
6O0V
 
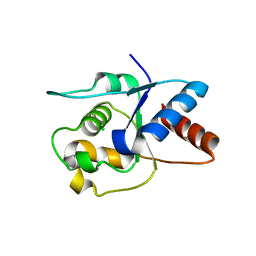 | | Crystal structure of the TIR domain G601P mutant from human SARM1, crystal form 2 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, Sterile alpha and TIR motif-containing protein 1 | | Authors: | Horsefield, S, Burdett, H, Zhang, X, Manik, M.K, Shi, Y, Chen, J, Tiancong, Q, Gilley, J, Lai, J, Gu, W, Rank, M, Casey, L, Ericsson, D.J, Foley, G, Hughes, R.O, Bosanac, T, von Itzstein, M, Rathjen, J.P, Nanson, J.D, Boden, M, Dry, I.B, Williams, S.J, Staskawicz, B.J, Coleman, M.P, Ve, T, Dodds, P.N, Kobe, B. | | Deposit date: | 2019-02-17 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | NAD+cleavage activity by animal and plant TIR domains in cell death pathways.
Science, 365, 2019
|
|
6O1B
 
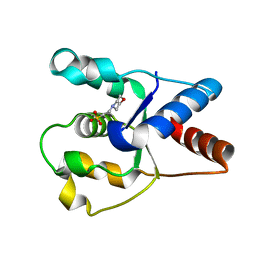 | | Crystal structure of the TIR domain G601P mutant from human SARM1, crystal form 1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Sterile alpha and TIR motif-containing protein 1 | | Authors: | Horsefield, S, Burdett, H, Zhang, X, Manik, M.K, Shi, Y, Chen, J, Tiancong, Q, Gilley, J, Lai, J, Gu, W, Rank, M, Casey, L, Ericsson, D.J, Foley, G, Hughes, R.O, Bosanac, T, von Itzstein, M, Rathjen, J.P, Nanson, J.D, Boden, M, Dry, I.B, Williams, S.J, Staskawicz, B.J, Coleman, M.P, Ve, T, Dodds, P.N, Kobe, B. | | Deposit date: | 2019-02-18 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | NAD+cleavage activity by animal and plant TIR domains in cell death pathways.
Science, 365, 2019
|
|
2RDD
 
 | | X-ray crystal structure of AcrB in complex with a novel transmembrane helix. | | Descriptor: | (2S,5R,6R)-6-{[(2R)-2-AMINO-2-PHENYLETHANOYL]AMINO}-3,3-DIMETHYL-7-OXO-4-THIA-1-AZABICYCLO[3.2.0]HEPTANE-2-CARBOXYLIC ACID, Acriflavine resistance protein B, UPF0092 membrane protein yajC | | Authors: | Tornroth-Horsefield, S, Gourdon, P, Horsefield, R, Neutze, R. | | Deposit date: | 2007-09-22 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of AcrB in complex with a single transmembrane subunit reveals another twist.
Structure, 15, 2007
|
|
5DYE
 
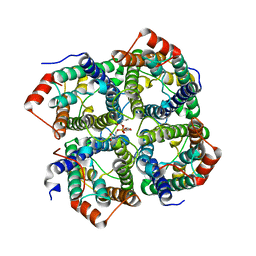 | | CRYSTAL STRUCTURE OF THE FULL LENGTH S156E MUTANT OF HUMAN AQUAPORIN 5 | | Descriptor: | Aquaporin-5, O-[(S)-{[(2S)-2-(hexanoyloxy)-3-(tetradecanoyloxy)propyl]oxy}(hydroxy)phosphoryl]-D-serine | | Authors: | Kitchen, P, Oeberg, F, Sjoehamn, J, Hedfalk, K, Bill, R.M, Conner, A.C, Conner, M.T, Toernroth-Horsefield, S. | | Deposit date: | 2015-09-24 | | Release date: | 2015-12-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Plasma Membrane Abundance of Human Aquaporin 5 Is Dynamically Regulated by Multiple Pathways.
Plos One, 10, 2015
|
|
5C5X
 
 | | CRYSTAL STRUCTURE OF THE S156E MUTANT OF HUMAN AQUAPORIN 5 | | Descriptor: | Aquaporin-5, O-[(S)-{[(2S)-2-(hexanoyloxy)-3-(tetradecanoyloxy)propyl]oxy}(hydroxy)phosphoryl]-D-serine | | Authors: | Kitchen, P, Oeberg, F, Sjoehamn, J, Hedfalk, K, Bill, R.M, Conner, A.C, Conner, M.T, Toernroth-Horsefield, S. | | Deposit date: | 2015-06-22 | | Release date: | 2015-12-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Plasma Membrane Abundance of Human Aquaporin 5 Is Dynamically Regulated by Multiple Pathways.
Plos One, 10, 2015
|
|
4IA4
 
 | |
3D9S
 
 | | Human Aquaporin 5 (AQP5) - High Resolution X-ray Structure | | Descriptor: | Aquaporin-5, O-[(S)-{[(2S)-2-(hexanoyloxy)-3-(tetradecanoyloxy)propyl]oxy}(hydroxy)phosphoryl]-D-serine | | Authors: | Horsefield, R, Norden, K, Fellert, M, Backmark, A, Tornroth-Horsefield, S, Terwisscha Van Scheltinga, A.C, Kvassman, J, Kjellbom, P, Johanson, U, Neutze, R. | | Deposit date: | 2008-05-27 | | Release date: | 2008-08-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution x-ray structure of human aquaporin 5
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
7W7R
 
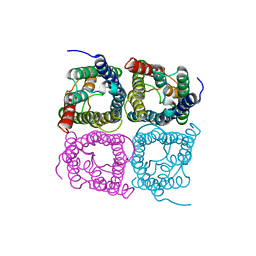 | | High resolution structure of a fish aquaporin reveals a novel extracellular fold. | | Descriptor: | Aquaporin 1 | | Authors: | Zeng, J, Schmitz, F, Isaksson, S, Glas, J, Arbab, O, Andersson, M, Sundell, K, Eriksson, L, Swaminathan, K, Tornroth-Horsefield, S, Hedfalk, K. | | Deposit date: | 2021-12-06 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | High-resolution structure of a fish aquaporin reveals a novel extracellular fold.
Life Sci Alliance, 5, 2022
|
|
7W7S
 
 | | High resolution structure of a fish aquaporin reveals a novel extracellular fold. | | Descriptor: | Aquaporin 1 | | Authors: | Zeng, J, Schmitz, F, Isaksson, S, Glas, J, Arbab, O, Andersson, M, Sundell, K, Eriksson, L, Swaminathan, K, Tornroth-Horsefield, S, Hedfalk, K. | | Deposit date: | 2021-12-06 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution structure of a fish aquaporin reveals a novel extracellular fold.
Life Sci Alliance, 5, 2022
|
|
2B5F
 
 | | Crystal structure of the spinach aquaporin SoPIP2;1 in an open conformation to 3.9 resolution | | Descriptor: | aquaporin | | Authors: | Tornroth-Horsefield, S, Wang, Y, Hedfalk, K, Johanson, U, Karlsson, M, Tajkhorshid, E, Neutze, R, Kjellbom, P. | | Deposit date: | 2005-09-28 | | Release date: | 2005-12-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structural mechanism of plant aquaporin gating
Nature, 439, 2006
|
|
4NEF
 
 | | X-ray structure of human Aquaporin 2 | | Descriptor: | Aquaporin-2, CADMIUM ION, ZINC ION | | Authors: | Frick, A, Eriksson, U, Mattia, F.D, Oberg, F, Hedfalk, K, Neutze, R, Grip, W.D, Deen, P.M.T, Tornroth-horsefield, S. | | Deposit date: | 2013-10-29 | | Release date: | 2014-04-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | X-ray structure of human aquaporin 2 and its implications for nephrogenic diabetes insipidus and trafficking
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1Z98
 
 | | Crystal structure of the spinach aquaporin SoPIP2;1 in a closed conformation | | Descriptor: | CADMIUM ION, aquaporin | | Authors: | Tornroth-Horsefield, S, Hedfalk, K, Johanson, U, Karlsson, M, Neutze, R, Kjellbom, P. | | Deposit date: | 2005-04-01 | | Release date: | 2005-12-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural mechanism of plant aquaporin gating
Nature, 439, 2006
|
|
4JC6
 
 | | Mercury activation of the plant aquaporin SoPIP2;1 - structural and functional characterization | | Descriptor: | Aquaporin, CADMIUM ION, MERCURY (II) ION, ... | | Authors: | Frick, A, Jarva, M, Nyblom, M, Ekvall, M, Uzdavinys, P, Tornroth-Horsefield, S. | | Deposit date: | 2013-02-21 | | Release date: | 2013-09-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.152 Å) | | Cite: | Mercury increases water permeability of a plant aquaporin through a non-cysteine-related mechanism.
Biochem.J., 454, 2013
|
|
3CLL
 
 | | Crystal structure of the Spinach Aquaporin SoPIP2;1 S115E mutant | | Descriptor: | Aquaporin | | Authors: | Nyblom, M, Alfredsson, A, Hallgren, K, Hedfalk, K, Neutze, R, Trnroth-Horsefield, S. | | Deposit date: | 2008-03-19 | | Release date: | 2009-02-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and functional analysis of SoPIP2;1 mutants adds insight into plant aquaporin gating.
J.Mol.Biol., 387, 2009
|
|
5UZB
 
 | | Cryo-EM structure of the MAL TIR domain filament | | Descriptor: | Toll/interleukin-1 receptor domain-containing adapter protein | | Authors: | Ve, T, Vajjhala, P.R, Hedger, A, Croll, T, DiMaio, F, Horsefield, S, Yu, X, Lavrencic, P, Hassan, Z, Morgan, G.P, Mansell, A, Mobli, M, O'Carrol, A, Chauvin, B, Gambin, Y, Sierecki, E, Landsberg, M.J, Stacey, K.J, Egelman, E.H, Kobe, B. | | Deposit date: | 2017-02-25 | | Release date: | 2017-07-26 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Structural basis of TIR-domain-assembly formation in MAL- and MyD88-dependent TLR4 signaling.
Nat. Struct. Mol. Biol., 24, 2017
|
|
3CN5
 
 | | Crystal structure of the Spinach Aquaporin SoPIP2;1 S115E, S274E mutant | | Descriptor: | Aquaporin | | Authors: | Nyblom, M, Alfredsson, A, Hallgren, K, Hedfalk, K, Neutze, R, Tornroth-Horsefield, S. | | Deposit date: | 2008-03-25 | | Release date: | 2009-02-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural and functional analysis of SoPIP2;1 mutants adds insight into plant aquaporin gating.
J.Mol.Biol., 387, 2009
|
|
3CN6
 
 | | Crystal structure of the Spinach Aquaporin SoPIP2;1 S274E mutant | | Descriptor: | Aquaporin, CADMIUM ION | | Authors: | Nyblom, M, Alfredsson, A, Hallgren, K, Hedfalk, K, Neutze, R, Tornroth-Horsefield, S. | | Deposit date: | 2008-03-25 | | Release date: | 2009-02-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural and functional analysis of SoPIP2;1 mutants adds insight into plant aquaporin gating.
J.Mol.Biol., 387, 2009
|
|
