2LNJ
 
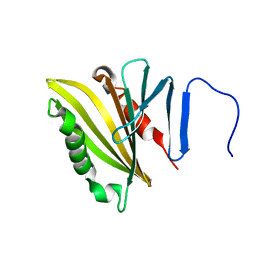 | |
2KMF
 
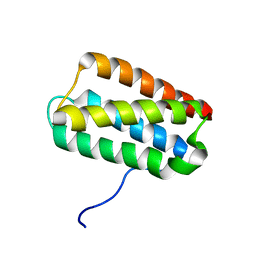 | | Solution Structure of Psb27 from cyanobacterial photosystem II | | Descriptor: | Photosystem II 11 kDa protein | | Authors: | Mabbitt, P.D, Rautureau, G.J.P, Day, C.L, Wilbanks, S.M, Eaton-Rye, J.J, Hinds, M.G. | | Deposit date: | 2009-07-28 | | Release date: | 2009-09-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Psb27 from cyanobacterial photosystem II
Biochemistry, 48, 2009
|
|
2JM6
 
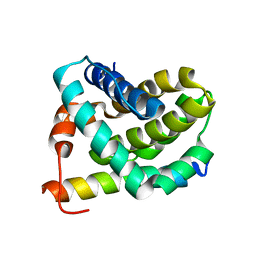 | | Solution structure of MCL-1 complexed with NOXAB | | Descriptor: | Myeloid cell leukemia-1 protein Mcl-1 homolog, Noxa | | Authors: | Czabotar, P.E, Lee, E.F, van Delft, M.F, Day, C.L, Smith, B.J, Huang, D.C.S, Fairlie, W.D, Hinds, M.G, Colman, P.M. | | Deposit date: | 2006-10-17 | | Release date: | 2007-03-20 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Structural insights into the degradation of Mcl-1 induced by BH3 domains
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2KUA
 
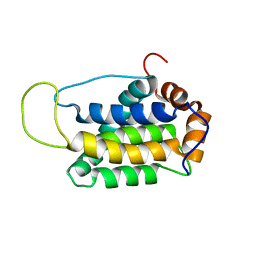 | |
1WSX
 
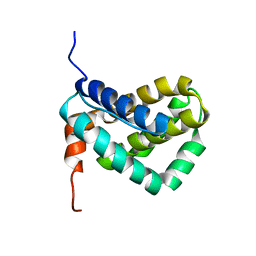 | | Solution structure of MCL-1 | | Descriptor: | myeloid cell leukemia sequence 1 | | Authors: | Day, C.L, Chen, L, Richardson, S.J, Harrison, P.J, Huang, D.C, Hinds, M.G. | | Deposit date: | 2004-11-12 | | Release date: | 2004-11-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Prosurvival Mcl-1 and Characterization of Its Binding by Proapoptotic BH3-only Ligands
J.Biol.Chem., 280, 2005
|
|
1ZEC
 
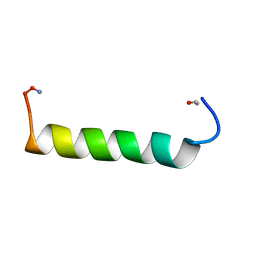 | | NMR Solution structure of NEF1-25, 20 structures | | Descriptor: | NEF1-25 | | Authors: | Barnham, K.J, Monks, S.A, Hinds, M.G, Azad, A.A, Norton, R.S. | | Deposit date: | 1996-12-18 | | Release date: | 1998-01-07 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a polypeptide from the N terminus of the HIV protein Nef.
Biochemistry, 36, 1997
|
|
1CWW
 
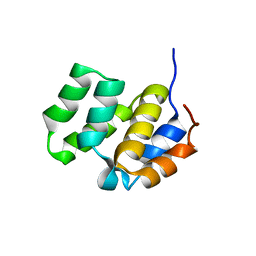 | | SOLUTION STRUCTURE OF THE CASPASE RECRUITMENT DOMAIN (CARD) FROM APAF-1 | | Descriptor: | APOPTOTIC PROTEASE ACTIVATING FACTOR 1 | | Authors: | Day, C.L, Dupont, C, Lackmann, M, Vaux, D.L, Hinds, M.G. | | Deposit date: | 1999-08-26 | | Release date: | 2000-01-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and mutagenesis of the caspase recruitment domain (CARD) from Apaf-1.
Cell Death Differ., 6, 1999
|
|
1RE6
 
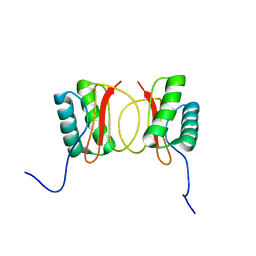 | | Localisation of Dynein Light Chains 1 and 2 and their Pro-apoptotic Ligands | | Descriptor: | dynein light chain 2 | | Authors: | Day, C.L, Puthalakath, H, Skea, G, Strasser, A, Barsukov, I, Lian, L.Y, Huang, D.C, Hinds, M.G. | | Deposit date: | 2003-11-06 | | Release date: | 2004-03-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Localization of dynein light chains 1 and 2 and their pro-apoptotic ligands.
Biochem.J., 377, 2004
|
|
1IXT
 
 | | Structure of a Novel P-Superfamily Spasmodic Conotoxin Reveals an Inhibitory Cystine Knot Motif | | Descriptor: | spasmodic protein tx9a-like protein | | Authors: | Miles, L.A, Dy, C.Y, Nielsen, J, Barnham, K.J, Hinds, M.G, Olivera, B.M, Bulaj, G, Norton, R.S. | | Deposit date: | 2002-07-04 | | Release date: | 2003-01-28 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of a Novel P-Superfamily Spasmodic Conotoxin Reveals an Inhibitory Cystine Knot Motif
J.Biol.Chem., 277, 2002
|
|
1OWT
 
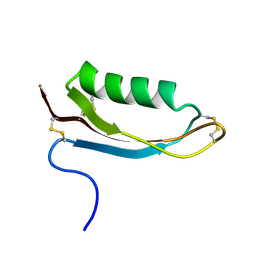 | | Structure of the Alzheimer's disease amyloid precursor protein copper binding domain | | Descriptor: | Amyloid beta A4 protein | | Authors: | Barnham, K.J, McKinstry, W.J, Multhaup, G, Galatis, D, Morton, C.J, Curtain, C.C, Williamson, N.A, White, A.R, Hinds, M.G, Norton, R.S, Beyreuther, K, Masters, C.L, Parker, M.W, Cappai, R. | | Deposit date: | 2003-03-30 | | Release date: | 2003-05-13 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structure of the Alzheimer's Disease Amyloid Precursor Protein Copper Binding Domain. A REGULATOR OF NEURONAL COPPER HOMEOSTASIS.
J.Biol.Chem., 278, 2003
|
|
7P0U
 
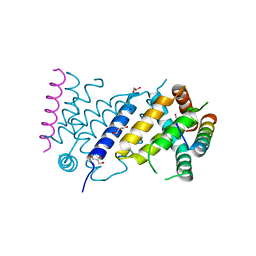 | | ORF virus encoded Bcl-2 homolog ORFV125 in complex with Puma BH3 peptide | | Descriptor: | 1,2-ETHANEDIOL, Activator of apoptosis harakiri, Apoptosis inhibitor, ... | | Authors: | Suraweera, C.D, Hinds, M.G, Kvansakul, M. | | Deposit date: | 2021-06-30 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.99374259 Å) | | Cite: | Structural Investigation of Orf Virus Bcl-2 Homolog ORFV125 Interactions with BH3-Motifs from BH3-Only Proteins Puma and Hrk.
Viruses, 13, 2021
|
|
7P0S
 
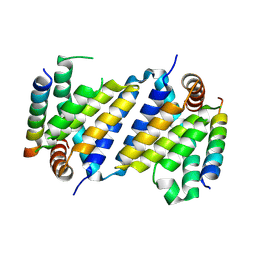 | |
7P33
 
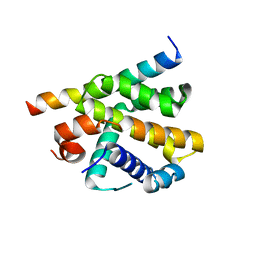 | | Epstein-Barr virus encoded Bcl-2 homolog BHRF-1 in complex with Bid BH3 peptide | | Descriptor: | 1,2-ETHANEDIOL, Apoptosis regulator BHRF1, BH3-interacting domain death agonist p15, ... | | Authors: | Suraweera, C.D, Hinds, M.G, Kvansakul, M. | | Deposit date: | 2021-07-07 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.78542733 Å) | | Cite: | Crystal Structures of Epstein-Barr Virus Bcl-2 Homolog BHRF1 Bound to Bid and Puma BH3 Motif Peptides.
Viruses, 14, 2022
|
|
7P9W
 
 | | Epstein-Barr virus encoded apoptosis regulator BHRF1 in complex with Puma BH3 | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, AMMONIUM ION, ... | | Authors: | Suraweera, C.D, Hinds, M.G, Kvansakul, M. | | Deposit date: | 2021-07-28 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.00010061 Å) | | Cite: | Crystal Structures of Epstein-Barr Virus Bcl-2 Homolog BHRF1 Bound to Bid and Puma BH3 Motif Peptides.
Viruses, 14, 2022
|
|
2ROC
 
 | | Solution structure of Mcl-1 Complexed with Puma | | Descriptor: | Bcl-2-binding component 3, Induced myeloid leukemia cell differentiation protein Mcl-1 homolog | | Authors: | Day, C.L, Smits, C, Fan, F.C, Lee, E.F, Fairlie, W.D, Hinds, M.G. | | Deposit date: | 2008-03-17 | | Release date: | 2008-07-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the BH3 Domains from the p53-Inducible BH3-Only Proteins Noxa and Puma in Complex with Mcl-1
J.Mol.Biol., 380, 2008
|
|
2ROD
 
 | | Solution Structure of MCL-1 Complexed with NoxaA | | Descriptor: | Induced myeloid leukemia cell differentiation protein Mcl-1 homolog, Noxa | | Authors: | Day, C.L, Smits, C, Fan, F.C, Lee, E.F, Fairlie, W.D, Hinds, M.G. | | Deposit date: | 2008-03-17 | | Release date: | 2008-07-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the BH3 Domains from the p53-Inducible BH3-Only Proteins Noxa and Puma in Complex with Mcl-1
J.Mol.Biol., 380, 2008
|
|
2VOI
 
 | | Structure of mouse A1 bound to the Bid BH3-domain | | Descriptor: | BCL-2-RELATED PROTEIN A1, BH3-INTERACTING DOMAIN DEATH AGONIST P13, CHLORIDE ION | | Authors: | Smits, C, Czabotar, P.E, Hinds, M.G, Day, C.L. | | Deposit date: | 2008-02-17 | | Release date: | 2008-03-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Plasticity Underpins Promiscuous Binding of the Prosurvival Protein A1.
Structure, 16, 2008
|
|
2VOG
 
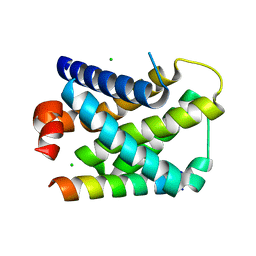 | | Structure of mouse A1 bound to the Bmf BH3-domain | | Descriptor: | BCL-2-MODIFYING FACTOR, BCL-2-RELATED PROTEIN A1, CHLORIDE ION | | Authors: | Smits, C, Czabotar, P.E, Hinds, M.G, Day, C.L. | | Deposit date: | 2008-02-17 | | Release date: | 2008-03-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Plasticity Underpins Promiscuous Binding of the Prosurvival Protein A1.
Structure, 16, 2008
|
|
2VOH
 
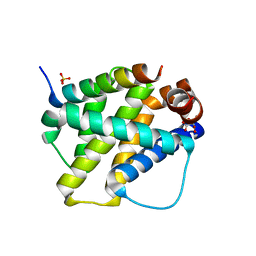 | | Structure of mouse A1 bound to the Bak BH3-domain | | Descriptor: | BCL-2 HOMOLOGOUS ANTAGONIST/KILLER, BCL-2-RELATED PROTEIN A1, CITRIC ACID, ... | | Authors: | Smits, C, Czabotar, P.E, Hinds, M.G, Day, C.L. | | Deposit date: | 2008-02-17 | | Release date: | 2008-03-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Plasticity Underpins Promiscuous Binding of the Prosurvival Protein A1.
Structure, 16, 2008
|
|
2VOF
 
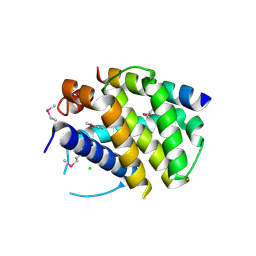 | | Structure of mouse A1 bound to the Puma BH3-domain | | Descriptor: | BCL-2-BINDING COMPONENT 3, BCL-2-RELATED PROTEIN A1, CHLORIDE ION | | Authors: | Smits, C, Czabotar, P.E, Hinds, M.G, Day, C.L. | | Deposit date: | 2008-02-17 | | Release date: | 2008-03-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Plasticity Underpins Promiscuous Binding of the Prosurvival Protein A1.
Structure, 16, 2008
|
|
