1KQN
 
 | | Crystal structure of NMN/NaMN adenylyltransferase complexed with NAD | | Descriptor: | NICOTINAMIDE MONONUCLEOTIDE ADENYLYL TRANSFERASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, XENON | | Authors: | Zhou, T, Kurnasov, O, Tomchick, D.R, Binns, D.D, Grishin, N.V, Marquez, V.E, Osterman, A.L, Zhang, H. | | Deposit date: | 2002-01-07 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Human Nicotinamide/Nicotonic Acid Mononucleotide Adenylyltransferase. Basis for the dual substrate specificity and activation of the oncolytic agent tiazofurin.
J.Biol.Chem., 277, 2003
|
|
4LNK
 
 | | B. subtilis glutamine synthetase structures reveal large active site conformational changes and basis for isoenzyme specific regulation: structure of GS-glutamate-AMPPCP complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLUTAMIC ACID, Glutamine synthetase, ... | | Authors: | Schumacher, M.A, Chinnam, N, Tonthat, N, Fisher, S, Wray, L. | | Deposit date: | 2013-07-11 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Structures of the Bacillus subtilis Glutamine Synthetase Dodecamer Reveal Large Intersubunit Catalytic Conformational Changes Linked to a Unique Feedback Inhibition Mechanism.
J.Biol.Chem., 288, 2013
|
|
1KR2
 
 | | CRYSTAL STRUCTURE OF HUMAN NMN/NAMN ADENYLYL TRANSFERASE COMPLEXED WITH TIAZOFURIN ADENINE DINUCLEOTIDE (TAD) | | Descriptor: | BETA-METHYLENE-THIAZOLE-4-CARBOXYAMIDE-ADENINE DINUCLEOTIDE, NICOTINAMIDE MONONUCLEOTIDE ADENYLYL TRANSFERASE | | Authors: | Zhou, T, Kurnasov, O, Tomchick, D.R, Binns, D.D, Grishin, N.V, Marquez, V.E, Osterman, A.L, Zhang, H. | | Deposit date: | 2002-01-08 | | Release date: | 2003-01-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Hhuman of Nicotinamide/Nicotinic Acid Mononucleotide Adenylyltransferase.
Basis for the dual substrate specificity and activation of the oncolytic agent tiazofurin.
J.Biol.Chem., 277, 2002
|
|
1KQO
 
 | | Crystal structure of NMN/NaMN adenylyltransferase complexed with deamido-NAD | | Descriptor: | NICOTINAMIDE MONONUCLEOTIDE ADENYLYL TRANSFERASE, NICOTINIC ACID ADENINE DINUCLEOTIDE | | Authors: | Zhou, T, Kurnasov, O, Tomchick, D.R, Binns, D.D, Grishin, N.V, Marquez, V.E, Osterman, A.L, Zhang, H. | | Deposit date: | 2002-01-07 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Hhuman of Nicotinamide/Nicotinic Acid Mononucleotide Adenylyltransferase.
Basis for the dual substrate specificity and activation of the oncolytic agent tiazofurin.
J.Biol.Chem., 277, 2002
|
|
1NUQ
 
 | | CRYSTAL STRUCTURE OF HUMAN CYTOSOLIC NMN/NaMN ADENYLYLTRANSFERASE COMPLEXED WITH NaAD | | Descriptor: | FKSG76, NICOTINIC ACID ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Zhang, X, Kurnasov, O.V, Karthikeyan, S, Grishin, N.V, Osterman, A.L, Zhang, H. | | Deposit date: | 2003-02-01 | | Release date: | 2003-06-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | STRUCTURAL CHARACTERIZATION OF A HUMAN CYTOSOLIC NMN/NaMN
ADENYLYLTRANSFERASE AND IMPLICATION IN HUMAN NAD BIOSYNTHESIS
J.Biol.Chem., 278, 2003
|
|
4LNF
 
 | | B. subtilis glutamine synthetase structures reveal large active site conformational changes and basis for isoenzyme specific regulation: structure of GS-Q | | Descriptor: | GLUTAMINE, Glutamine synthetase, MAGNESIUM ION, ... | | Authors: | Schumacher, M.A, Chinnam, N, Tonthat, N, Fisher, S, Wray, L. | | Deposit date: | 2013-07-11 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.949 Å) | | Cite: | Structures of the Bacillus subtilis Glutamine Synthetase Dodecamer Reveal Large Intersubunit Catalytic Conformational Changes Linked to a Unique Feedback Inhibition Mechanism.
J.Biol.Chem., 288, 2013
|
|
4LNN
 
 | | B. subtilis glutamine synthetase structures reveal large active site conformational changes and basis for isoenzyme specific regulation: structure of apo form of GS | | Descriptor: | Glutamine synthetase, MAGNESIUM ION, SULFATE ION | | Authors: | Schumacher, M.A, Chinnam, N, Tonthat, N, Fisher, S, Wray, L. | | Deposit date: | 2013-07-11 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of the Bacillus subtilis Glutamine Synthetase Dodecamer Reveal Large Intersubunit Catalytic Conformational Changes Linked to a Unique Feedback Inhibition Mechanism.
J.Biol.Chem., 288, 2013
|
|
4LNO
 
 | | B. subtilis glutamine synthetase structures reveal large active site conformational changes and basis for isoenzyme specific regulation: form two of GS-1 | | Descriptor: | GLUTAMINE, Glutamine synthetase, MAGNESIUM ION | | Authors: | Schumacher, M.A, Chinnam, N, Tonthat, N, Fisher, S, Wray, L. | | Deposit date: | 2013-07-11 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of the Bacillus subtilis Glutamine Synthetase Dodecamer Reveal Large Intersubunit Catalytic Conformational Changes Linked to a Unique Feedback Inhibition Mechanism.
J.Biol.Chem., 288, 2013
|
|
4LNI
 
 | | B. subtilis glutamine synthetase structures reveal large active site conformational changes and basis for isoenzyme specific regulation: structure of the transition state complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Glutamine synthetase, L-METHIONINE-S-SULFOXIMINE PHOSPHATE, ... | | Authors: | Schumacher, M.A, Chinnam, N, Tonthat, N, Fisher, S, Wray, L. | | Deposit date: | 2013-07-11 | | Release date: | 2013-11-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5793 Å) | | Cite: | Structures of the Bacillus subtilis Glutamine Synthetase Dodecamer Reveal Large Intersubunit Catalytic Conformational Changes Linked to a Unique Feedback Inhibition Mechanism.
J.Biol.Chem., 288, 2013
|
|
1NUR
 
 | | CRYSTAL STRUCTURE OF HUMAN CYTOSOLIC NMN/NaMN ADENYLYLTRANSFERASE | | Descriptor: | FKSG76, SULFATE ION | | Authors: | Zhang, X, Kurnasov, O.V, Karthikeyan, S, Grishin, N.V, Osterman, A.L, Zhang, H. | | Deposit date: | 2003-02-01 | | Release date: | 2003-06-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | STRUCTURAL CHARACTERIZATION OF A HUMAN CYTOSOLIC NMN/NaMN
ADENYLYLTRANSFERASE AND IMPLICATION IN HUMAN NAD BIOSYNTHESIS
J.Biol.Chem., 278, 2003
|
|
3AH9
 
 | | Crystal structure of (Pro-Pro-Gly)9 at 1.1 A resolution | | Descriptor: | ACETIC ACID, AZIDE ION, collagen-like peptide | | Authors: | Okuyama, K, Morimoto, T, Hongo, C, Hosaka, N, Nishino, N. | | Deposit date: | 2010-04-22 | | Release date: | 2011-04-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Crystal structure of (Pro-Pro-Gly)9
To be Published
|
|
2HWR
 
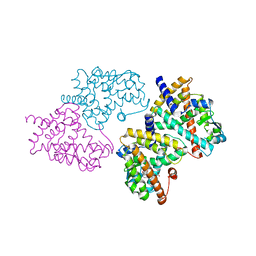 | | Structural basis for the structure-activity relationships of Peroxisome Proliferator-Activated Receptor agonists | | Descriptor: | 2-[(1-{3-[(6-BENZOYL-1-PROPYL-2-NAPHTHYL)OXY]PROPYL}-1H-INDOL-4-YL)OXY]-2-METHYLPROPANOIC ACID, Peroxisome proliferator-activated receptor gamma | | Authors: | Peng, Y.H, Lu, I.L, Mahindroo, N, Lin, C.H, Hsieh, H.P, Wu, S.Y. | | Deposit date: | 2006-08-01 | | Release date: | 2007-08-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural basis for the structure-activity relationships of peroxisome proliferator-activated receptor agonists
J.Med.Chem., 49, 2006
|
|
2HWQ
 
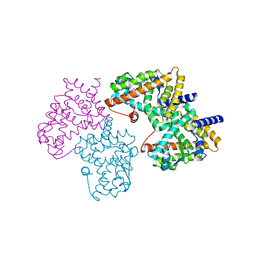 | | Structural basis for the structure-activity relationships of Peroxisome Proliferator-Activated Receptor agonists | | Descriptor: | Peroxisome proliferator-activated receptor gamma, [(1-{3-[(6-BENZOYL-1-PROPYL-2-NAPHTHYL)OXY]PROPYL}-1H-INDOL-5-YL)OXY]ACETIC ACID | | Authors: | Peng, Y.H, Lu, I.L, Mahindroo, N, Lin, C.H, Hsieh, H.P, Wu, S.Y. | | Deposit date: | 2006-08-01 | | Release date: | 2007-08-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural basis for the structure-activity relationships of peroxisome proliferator-activated receptor agonists
J.Med.Chem., 49, 2006
|
|
1NB0
 
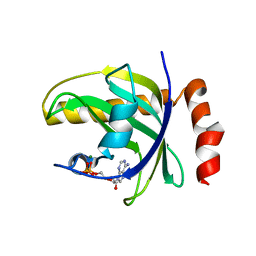 | | Crystal Structure of Human Riboflavin Kinase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, hypothetical protein FLJ11149 | | Authors: | Karthikeyan, S, Zhou, Q, Mseeh, F, Grishin, N.V, Osterman, A.L, Zhang, H. | | Deposit date: | 2002-12-01 | | Release date: | 2003-03-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Human Riboflavin Kinase Reveals a Beta Barrel Fold and a Novel Active Site Arch
Structure, 11, 2003
|
|
4LSD
 
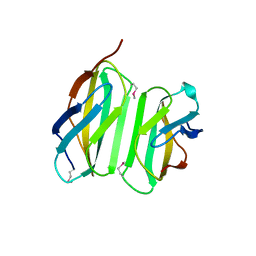 | | Myokine structure | | Descriptor: | Fibronectin type III domain-containing protein 5 | | Authors: | Schumacher, M.A, Ohashi, T, Shah, R.S, Chinnam, N, Erickson, H. | | Deposit date: | 2013-07-22 | | Release date: | 2013-10-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | The structure of irisin reveals a novel intersubunit beta-sheet fibronectin type III (FNIII) dimer: implications for receptor activation.
J.Biol.Chem., 288, 2013
|
|
1LW7
 
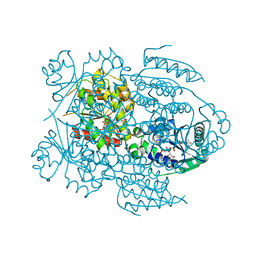 | | NADR PROTEIN FROM HAEMOPHILUS INFLUENZAE | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, TRANSCRIPTIONAL REGULATOR NADR | | Authors: | Singh, S.K, Kurnasov, O.V, Chen, B, Robinson, H, Grishin, N.V, Osterman, A.L, Zhang, H. | | Deposit date: | 2002-05-30 | | Release date: | 2002-08-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of Haemophilus influenzae NadR protein. A bifunctional enzyme endowed with NMN adenyltransferase and ribosylnicotinimide kinase activities.
J.Biol.Chem., 277, 2002
|
|
2KBG
 
 | | Solution structure of the second Fibronectin type-III module of NCAM2 | | Descriptor: | Neural cell adhesion molecule 2 | | Authors: | Rasmussen, K.K, Teilum, K, Kulahin, N, Clausen, O, Berezin, V, Bock, E, Walmod, P.S, Poulsen, F.M. | | Deposit date: | 2008-11-28 | | Release date: | 2009-12-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the second Fibronectin type-III module of NCAM2
To be Published
|
|
3AJI
 
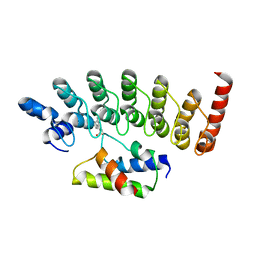 | | Structure of Gankyrin-S6ATPase photo-cross-linked site-specifically, and incoporated by genetic code expansion | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 10, Proteasome (Prosome, macropain) 26S subunit, ... | | Authors: | Sato, S, Mimasu, S, Sato, A, Hino, N, Sakamoto, K, Umehara, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-06-07 | | Release date: | 2010-12-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystallographic study of a site-specifically cross-linked protein complex with a genetically incorporated photoreactive amino acid
Biochemistry, 50, 2011
|
|
1V9J
 
 | | Solution structure of a BolA-like protein from Mus musculus | | Descriptor: | BolA-like protein RIKEN cDNA 1110025L05 | | Authors: | Kasai, T, Inoue, M, Koshiba, S, Yabuki, T, Aoki, M, Nunokawa, E, Seki, E, Matsuda, T, Matsuda, N, Tomo, Y, Shirouzu, M, Terada, T, Obayashi, N, Hamana, H, Shinya, N, Tatsuguchi, A, Yasuda, S, Yoshida, M, Hirota, H, Matsuo, Y, Tani, K, Suzuki, H, Arakawa, T, Carninci, P, Kawai, J, Hayashizaki, Y, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-26 | | Release date: | 2004-02-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a BolA-like protein from Mus musculus
Protein Sci., 13, 2004
|
|
2KDO
 
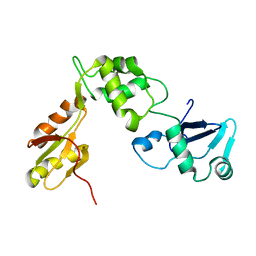 | | Structure of the human Shwachman-Bodian-Diamond syndrome protein, SBDS | | Descriptor: | Ribosome maturation protein SBDS | | Authors: | de Oliveira, J.F, Sforca, M.L, Blumenschein, T, Guimaraes, B.G, Zanchin, N.I.T, Zeri, A.C. | | Deposit date: | 2009-01-14 | | Release date: | 2010-01-19 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structure, dynamics, and RNA interaction analysis of the human SBDS protein.
J.Mol.Biol., 396, 2010
|
|
3WNO
 
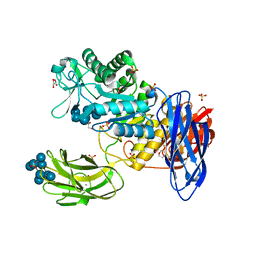 | | D308A mutant of Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase complexed with cycloisomaltooctaose | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Suzuki, N, Fujimoto, Z, Kim, Y.M, Momma, M, Kishine, N, Suzuki, R, Kobayashi, M, Kimura, A, Funane, K. | | Deposit date: | 2013-12-10 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural elucidation of the cyclization mechanism of alpha-1,6-glucan by Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase.
J.Biol.Chem., 289, 2014
|
|
3WNM
 
 | | D308A mutant of Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase complexed with isomaltoheptaose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Cycloisomaltooligosaccharide glucanotransferase, ... | | Authors: | Suzuki, N, Fujimoto, Z, Kim, Y.M, Momma, M, Kishine, N, Suzuki, R, Suzuki, S, Kitamura, S, Kobayashi, M, Kimura, A, Funane, K. | | Deposit date: | 2013-12-10 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural elucidation of the cyclization mechanism of alpha-1,6-glucan by Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase.
J.Biol.Chem., 289, 2014
|
|
3WNL
 
 | | D308A mutant of Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase complexed with isomaltohexaose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Cycloisomaltooligosaccharide glucanotransferase, ... | | Authors: | Suzuki, N, Fujimoto, Z, Kim, Y.M, Momma, M, Kishine, N, Suzuki, R, Suzuki, S, Kitamura, S, Kobayashi, M, Kimura, A, Funane, K. | | Deposit date: | 2013-12-10 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural elucidation of the cyclization mechanism of alpha-1,6-glucan by Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase.
J.Biol.Chem., 289, 2014
|
|
2P38
 
 | |
3WNK
 
 | | Crystal Structure of Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase | | Descriptor: | ACETATE ION, CADMIUM ION, CALCIUM ION, ... | | Authors: | Suzuki, N, Fujimoto, Z, Kim, Y.M, Momma, M, Kishine, N, Suzuki, R, Kobayashi, M, Kimura, A, Funane, K. | | Deposit date: | 2013-12-10 | | Release date: | 2014-02-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural elucidation of the cyclization mechanism of alpha-1,6-glucan by Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase.
J.Biol.Chem., 289, 2014
|
|
