1RZK
 
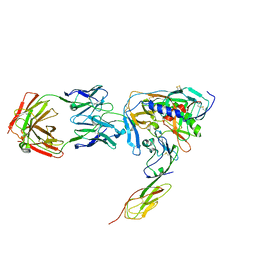 | | HIV-1 YU2 GP120 ENVELOPE GLYCOPROTEIN COMPLEXED WITH CD4 AND INDUCED NEUTRALIZING ANTIBODY 17B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ANTIBODY 17B, HEAVY CHAIN, ... | | Authors: | Huang, C.C, Venturi, M, Majeed, S, Moore, M.J, Phogat, S, Zhang, M.-Y, Dimitrov, D.S, Hendrickson, W.A, Robinson, J, Sodroski, J, Wyatt, R, Choe, H, Farzan, M, Kwong, P.D. | | Deposit date: | 2003-12-24 | | Release date: | 2004-02-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of tyrosine sulfation and VH-gene usage in antibodies that recognize the HIV type 1 coreceptor-binding site on gp120
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1AY9
 
 | | WILD-TYPE UMUD' FROM E. COLI | | Descriptor: | UMUD PROTEIN | | Authors: | Peat, T.S, Frank, E.G, Mcdonald, J.P, Levine, A.S, Woodgate, R, Hendrickson, W.A. | | Deposit date: | 1997-11-15 | | Release date: | 1998-01-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The UmuD' protein filament and its potential role in damage induced mutagenesis.
Structure, 4, 1996
|
|
1AV5
 
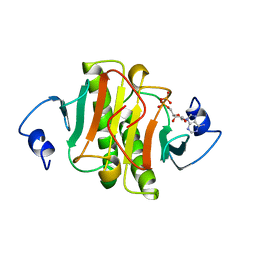 | | PKCI-SUBSTRATE ANALOG | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, PROTEIN KINASE C INTERACTING PROTEIN | | Authors: | Lima, C.D, Klein, M.G, Hendrickson, W.A. | | Deposit date: | 1997-09-25 | | Release date: | 1998-03-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based analysis of catalysis and substrate definition in the HIT protein family.
Science, 278, 1997
|
|
3LIF
 
 | |
3LIE
 
 | |
3LIC
 
 | |
3LI9
 
 | |
3LCK
 
 | |
3LI8
 
 | |
3LIA
 
 | |
1HR3
 
 | |
7SIM
 
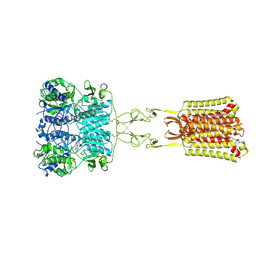 | | Structure of positive allosteric modulator-free active human calcium-sensing receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Park, J, Zuo, H, Frangaj, A, Fu, Z, Yen, L.Y, Zhang, Z, Mosyak, L, Slavkovich, V.N, Liu, J, Ray, K.M, Cao, B, Vallese, F, Geng, Y, Chen, S, Grassucci, R, Dandey, V.P, Tan, Y.Z, Eng, E, Lee, Y, Kloss, B, Liu, Z, Hendrickson, W.A, Potter, C.S, Carragher, B, Graziano, J, Conigrave, A.D, Frank, J, Clarke, O.B, Fan, Q.R. | | Deposit date: | 2021-10-14 | | Release date: | 2022-01-19 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Symmetric activation and modulation of the human calcium-sensing receptor.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
3M78
 
 | |
7SIL
 
 | | Structure of positive allosteric modulator-bound active human calcium-sensing receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(2-chlorophenyl)-N-[(1R)-1-(3-methoxyphenyl)ethyl]propan-1-amine, ... | | Authors: | Park, J, Zuo, H, Frangaj, A, Fu, Z, Yen, L.Y, Zhang, Z, Mosyak, L, Slavkovich, V.N, Liu, J, Ray, K.M, Cao, B, Vallese, F, Geng, Y, Chen, S, Grassucci, R, Dandey, V.P, Tan, Y.Z, Eng, E, Lee, Y, Kloss, B, Liu, Z, Hendrickson, W.A, Potter, C.S, Carragher, B, Graziano, J, Conigrave, A.D, Frank, J, Clarke, O.B, Fan, Q.R. | | Deposit date: | 2021-10-14 | | Release date: | 2022-01-19 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Symmetric activation and modulation of the human calcium-sensing receptor.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
3M70
 
 | |
3M75
 
 | |
3M72
 
 | |
3M74
 
 | |
3M7E
 
 | |
3M76
 
 | |
3M73
 
 | |
3M7B
 
 | |
3LID
 
 | |
3M7L
 
 | |
3M77
 
 | |
