7MMO
 
 | | LY-CoV1404 neutralizing antibody against SARS-CoV-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LY-CoV1404 Fab heavy chain, LY-CoV1404 Fab light chain, ... | | Authors: | Hendle, J, Pustilnik, A, Sauder, J.M, Coleman, K.A, Boyles, J.S, Dickinson, C.D. | | Deposit date: | 2021-04-30 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.427 Å) | | Cite: | LY-CoV1404 (bebtelovimab) potently neutralizes SARS-CoV-2 variants.
Biorxiv, 2022
|
|
6MNH
 
 | | ULK1 Unc-51 like autophagy activating kinase in complex with inhibitor BTC | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, N-[(2R)-3-methylbutan-2-yl]-1H-benzotriazole-6-carboxamide, ... | | Authors: | Hendle, J, Sauder, J.M, Hickey, M.J, Rauch, C.T, Maletic, M, Schwinn, K.D. | | Deposit date: | 2018-10-01 | | Release date: | 2019-03-27 | | Last modified: | 2019-11-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Idea2Data: Toward a New Paradigm for Drug Discovery.
Acs Med.Chem.Lett., 10, 2019
|
|
8F60
 
 | | anti-BTLA monoclonal antibody r23C8 in complex with BTLA | | Descriptor: | ACETATE ION, B- and T-lymphocyte attenuator, alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hendle, J, Atwell, S, Lieu, R, Hickey, M, Weichert, K. | | Deposit date: | 2022-11-15 | | Release date: | 2023-05-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Epitope topography of agonist antibodies to the checkpoint inhibitory receptor BTLA.
Structure, 31, 2023
|
|
8F6O
 
 | | anti-BTLA monoclonal antibody h22B3 in complex with BTLA | | Descriptor: | B- and T-lymphocyte attenuator, h22B3 Fab heavy chain, h22B3 Fab light chain | | Authors: | Hendle, J, Atwell, S, Lieu, R, Hickey, M, Weichert, K. | | Deposit date: | 2022-11-16 | | Release date: | 2023-05-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Epitope topography of agonist antibodies to the checkpoint inhibitory receptor BTLA.
Structure, 31, 2023
|
|
8F6L
 
 | | anti-BTLA monoclonal antibody h25F7 in complex with BTLA | | Descriptor: | B- and T-lymphocyte attenuator, h25F7 Fab heavy chain, h25F7 Fab light chain | | Authors: | Hendle, J, Atwell, S, Lieu, R, Hickey, M, Weichert, K. | | Deposit date: | 2022-11-16 | | Release date: | 2023-05-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Epitope topography of agonist antibodies to the checkpoint inhibitory receptor BTLA.
Structure, 31, 2023
|
|
8V9B
 
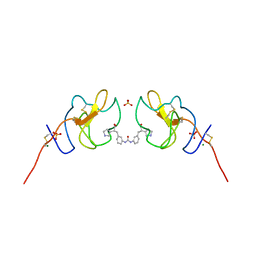 | | Lipoprotein(a) Kringle IV domain 7 - Lp(a) KIV7 in complex with LY3441732 | | Descriptor: | (2S,2'S)-3,3'-[carbonylbis(azanediyl-3,1-phenylene)]bis{2-[(3R)-pyrrolidin-1-ium-3-yl]propanoate}, Apolipoprotein(a), MAGNESIUM ION, ... | | Authors: | Hendle, J, Weichert, K, Sauder, J.M. | | Deposit date: | 2023-12-07 | | Release date: | 2024-05-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Discovery of potent small-molecule inhibitors of lipoprotein(a) formation.
Nature, 629, 2024
|
|
8TCE
 
 | |
8V8Z
 
 | |
2WB0
 
 | |
2WAZ
 
 | |
7MYR
 
 | | BACE-1 in complex with compound #18 | | Descriptor: | (4aR,7aR)-6-(5-fluoropyrimidin-2-yl)-7a-(1,2-thiazol-5-yl)-4,4a,5,6,7,7a-hexahydropyrrolo[3,4-d][1,3]thiazin-2-amine, Beta-secretase 1, GLYCEROL, ... | | Authors: | Hendle, J, Timm, D.E, Stout, S.L. | | Deposit date: | 2021-05-21 | | Release date: | 2021-07-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery and Early Clinical Development of LY3202626, a Low-Dose, CNS-Penetrant BACE Inhibitor.
J.Med.Chem., 64, 2021
|
|
7MYU
 
 | | BACE-1 in complex with compound #22 | | Descriptor: | Beta-secretase 1, N-{3-[(4aR,7aS)-2-amino-6-(5-fluoropyrimidin-2-yl)-4a,5,6,7-tetrahydropyrrolo[3,4-d][1,3]thiazin-7a(4H)-yl]-4-fluorophenyl}-5-methoxypyrazine-2-carboxamide, SULFATE ION | | Authors: | Hendle, J, Timm, D.E, Stout, S.L. | | Deposit date: | 2021-05-21 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Discovery and Early Clinical Development of LY3202626, a Low-Dose, CNS-Penetrant BACE Inhibitor.
J.Med.Chem., 64, 2021
|
|
7MYI
 
 | | BACE-1 in complex with compound #6 | | Descriptor: | (4aR,7aR)-6-(pyrimidin-2-yl)-7a-(thiophen-2-yl)-4,4a,5,6,7,7a-hexahydropyrrolo[3,4-d][1,3]thiazin-2-amine, Beta-secretase 1, GLYCEROL | | Authors: | Hendle, J, Timm, D.E, Stout, S.L. | | Deposit date: | 2021-05-21 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Discovery and Early Clinical Development of LY3202626, a Low-Dose, CNS-Penetrant BACE Inhibitor.
J.Med.Chem., 64, 2021
|
|
5VR9
 
 | | CH1/Ckappa Fab based on Matuzumab | | Descriptor: | CH1/Ckappa Fab heavy chain, CH1/Ckappa Fab light chain | | Authors: | Hendle, J. | | Deposit date: | 2017-05-10 | | Release date: | 2017-08-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Computational design of a specific heavy chain/ kappa light chain interface for expressing fully IgG bispecific antibodies.
Protein Sci., 26, 2017
|
|
5VSH
 
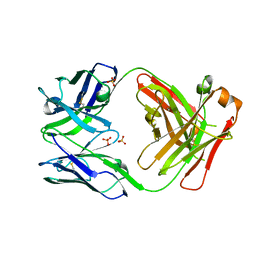 | | CH1/Clambda Fab based on Pertuzumab | | Descriptor: | CH1/Clambda Fab heavy chain, CH1/Clambda Fab light chain, SULFATE ION | | Authors: | Hendle, J. | | Deposit date: | 2017-05-11 | | Release date: | 2017-08-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Computational design of a specific heavy chain/ kappa light chain interface for expressing fully IgG bispecific antibodies.
Protein Sci., 26, 2017
|
|
5VSI
 
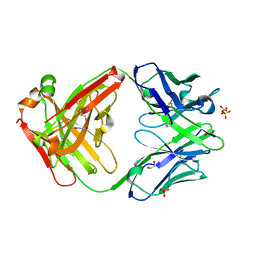 | | CH1/Ckappa Fab mutant 15.1 | | Descriptor: | 1,2-ETHANEDIOL, CH1/Ckappa Fab heavy chain, CH1/Ckappa Fab light chain, ... | | Authors: | Hendle, J. | | Deposit date: | 2017-05-11 | | Release date: | 2017-08-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Computational design of a specific heavy chain/ kappa light chain interface for expressing fully IgG bispecific antibodies.
Protein Sci., 26, 2017
|
|
7KMG
 
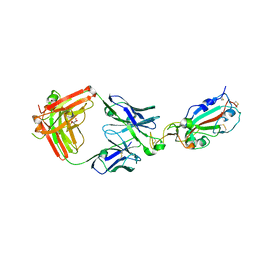 | | LY-CoV555 neutralizing antibody against SARS-CoV-2 | | Descriptor: | GLYCEROL, LY-CoV555 Fab heavy chain, LY-CoV555 Fab light chain, ... | | Authors: | Hendle, J, Pustilnik, A, Sauder, J.M, Coleman, K.A, Boyles, J.S, Dickinson, C.D. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | The neutralizing antibody, LY-CoV555, protects against SARS-CoV-2 infection in nonhuman primates.
Sci Transl Med, 13, 2021
|
|
7KMH
 
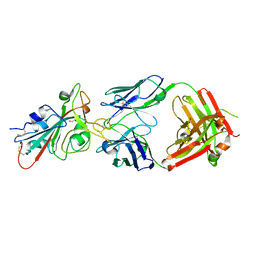 | | LY-CoV488 neutralizing antibody against SARS-CoV-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, LY-CoV488 Fab heavy chain, ... | | Authors: | Hendle, J, Pustilnik, A, Sauder, J.M, Boyles, J.S, Dickinson, C.D, Coleman, K.A. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The neutralizing antibody, LY-CoV555, protects against SARS-CoV-2 infection in nonhuman primates.
Sci Transl Med, 13, 2021
|
|
7KMI
 
 | | LY-CoV481 neutralizing antibody against SARS-CoV-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, LY-CoV481 Fab heavy chain, ... | | Authors: | Hendle, J, Pustilnik, A, Sauder, J.M, Coleman, K.A, Boyles, J.S, Dickinson, C.D. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | The neutralizing antibody, LY-CoV555, protects against SARS-CoV-2 infection in nonhuman primates.
Sci Transl Med, 13, 2021
|
|
1DPD
 
 | |
1DPC
 
 | |
1DPB
 
 | |
1GPE
 
 | | GLUCOSE OXIDASE FROM PENICILLIUM AMAGASAKIENSE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Hendle, J, Kalisz, H.M, Hecht, H.J. | | Deposit date: | 1999-03-24 | | Release date: | 1999-05-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 and 1.9 A resolution structures of the Penicillium amagasakiense and Aspergillus niger glucose oxidases as a basis for modelling substrate complexes.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
6UWP
 
 | | BACE-1 in complex with compound #32 | | Descriptor: | (1R,2R)-2-[(4aR,7aR)-2-amino-6-(pyrimidin-2-yl)-4a,5,6,7-tetrahydropyrrolo[3,4-d][1,3]thiazin-7a(4H)-yl]-N-{[(1R,2R)-2-methylcyclopropyl]methyl}cyclopropane-1-carboxamide, Beta-secretase 1, GLYCEROL, ... | | Authors: | Hendle, J, Timm, D.E. | | Deposit date: | 2019-11-05 | | Release date: | 2019-12-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Preparation and biological evaluation of BACE1 inhibitors: Leveraging trans-cyclopropyl moieties as ligand efficient conformational constraints.
Bioorg.Med.Chem., 28, 2020
|
|
6UVP
 
 | | BACE-1 in complex with compound #3 | | Descriptor: | Beta-secretase 1, GLYCEROL, N-{(1R,2R)-2-[(4aS,7aR)-2-amino-4a,5-dihydro-4H-furo[3,4-d][1,3]thiazin-7a(7H)-yl]cyclopropyl}-5-fluoropyridine-2-carboxamide, ... | | Authors: | Hendle, J, Timm, D.E. | | Deposit date: | 2019-11-04 | | Release date: | 2019-12-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Preparation and biological evaluation of BACE1 inhibitors: Leveraging trans-cyclopropyl moieties as ligand efficient conformational constraints.
Bioorg.Med.Chem., 28, 2020
|
|
