4KB5
 
 | | Crystal structure of MycP1 from Mycobacterium smegmatis | | Descriptor: | GLYCEROL, Membrane-anchored mycosin mycp1 | | Authors: | Sun, D.M, He, Y, Tian, C.L. | | Deposit date: | 2013-04-23 | | Release date: | 2014-02-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The putative propeptide of MycP1 in mycobacterial type VII secretion system does not inhibit protease activity but improves protein stability.
Protein Cell, 4, 2013
|
|
4M1Z
 
 | | Crystal structure of MycP1 with the N-terminal propeptide removed | | Descriptor: | Membrane-anchored mycosin mycp1 | | Authors: | Sun, D.M, He, Y, Wang, C.L, Zang, J.Y, Tian, C.L. | | Deposit date: | 2013-08-04 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The putative propeptide of MycP1 in mycobacterial type VII secretion system does not inhibit protease activity but improves protein stability.
Protein Cell, 4, 2013
|
|
5FUR
 
 | | Structure of human TFIID-IIA bound to core promoter DNA | | Descriptor: | SUPER CORE PROMOTER, TATA-BOX-BINDING PROTEIN, TRANSCRIPTION INITIATION FACTOR IIA SUBUNIT 1, ... | | Authors: | Louder, R.K, He, Y, Lopez-Blanco, J.R, Fang, J, Chacon, P, Nogales, E. | | Deposit date: | 2016-01-29 | | Release date: | 2016-04-06 | | Last modified: | 2017-08-02 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | Structure of Promoter-Bound TFIID and Model of Human Pre-Initiation Complex Assembly.
Nature, 531, 2016
|
|
5XX0
 
 | |
5XWX
 
 | |
7KJS
 
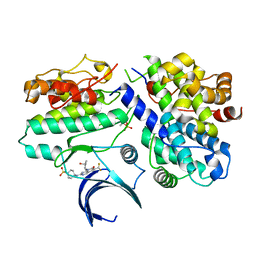 | | Crystal structure of CDK2/cyclin E in complex with PF-06873600 | | Descriptor: | 6-(difluoromethyl)-8-[(1R,2R)-2-hydroxy-2-methylcyclopentyl]-2-{[1-(methylsulfonyl)piperidin-4-yl]amino}pyrido[2,3-d]pyrimidin-7(8H)-one, Cyclin-dependent kinase 2, G1/S-specific cyclin-E1 | | Authors: | McTigue, M.A, He, Y, Ferre, R.A. | | Deposit date: | 2020-10-26 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.187 Å) | | Cite: | Discovery of PF-06873600, a CDK2/4/6 Inhibitor for the Treatment of Cancer.
J.Med.Chem., 64, 2021
|
|
5YR0
 
 | | Structure of Beclin1-UVRAG coiled coil domain complex | | Descriptor: | Beclin-1, UV radiation resistance associated protein | | Authors: | Pan, X, Zhao, Y, He, Y. | | Deposit date: | 2017-11-08 | | Release date: | 2018-06-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Targeting the potent Beclin 1-UVRAG coiled-coil interaction with designed peptides enhances autophagy and endolysosomal trafficking.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8HNA
 
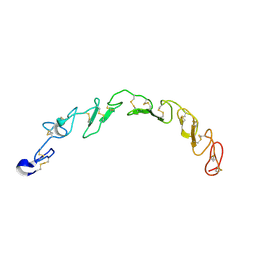 | |
8HN0
 
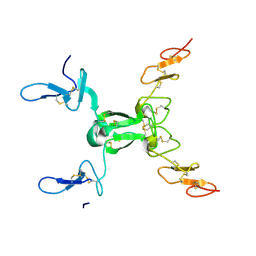 | |
1YC1
 
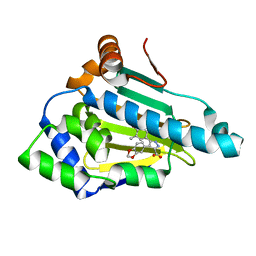 | | Crystal Structures of human HSP90alpha complexed with dihydroxyphenylpyrazoles | | Descriptor: | 4-(1,3-BENZODIOXOL-5-YL)-5-(5-ETHYL-2,4-DIHYDROXYPHENYL)-2H-PYRAZOLE-3-CARBOXYLIC ACID, Heat shock protein HSP 90-alpha | | Authors: | Kreusch, A, Han, S, Brinker, A, Zhou, V, Choi, H, He, Y, Lesley, S.A, Caldwell, J, Gu, X. | | Deposit date: | 2004-12-21 | | Release date: | 2005-02-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of human HSP90alpha-complexed with dihydroxyphenylpyrazoles.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
1YC4
 
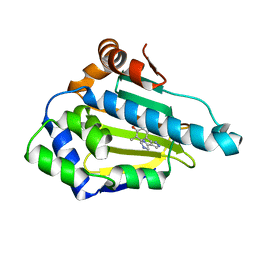 | | Crystal structure of human HSP90alpha complexed with dihydroxyphenylpyrazoles | | Descriptor: | 4-(1H-IMIDAZOL-4-YL)-3-(5-ETHYL-2,4-DIHYDROXY-PHENYL)-1H-PYRAZOLE, Heat shock protein HSP 90-alpha | | Authors: | Kreusch, A, Han, S, Brinker, A, Zhou, V, Choi, H, He, Y, Lesley, S.A, Caldwell, J, Gu, X. | | Deposit date: | 2004-12-21 | | Release date: | 2005-02-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structures of human HSP90alpha-complexed with dihydroxyphenylpyrazoles.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
1YC3
 
 | | Crystal Structure of human HSP90alpha complexed with dihydroxyphenylpyrazoles | | Descriptor: | 4-(1,3-BENZODIOXOL-5-YL)-5-(5-ETHYL-2,4-DIHYDROXYPHENYL)-2H-PYRAZOLE-3-CARBOXYLIC ACID, Heat shock protein HSP 90-alpha | | Authors: | Kreusch, A, Han, S, Brinker, A, Zhou, V, Choi, H, He, Y, Lesley, S.A, Caldwell, J, Gu, X. | | Deposit date: | 2004-12-21 | | Release date: | 2005-02-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal structures of human HSP90alpha-complexed with dihydroxyphenylpyrazoles.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
2OUT
 
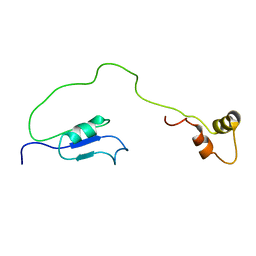 | | Solution Structure of HI1506, a Novel Two Domain Protein from Haemophilus influenzae | | Descriptor: | Mu-like prophage FluMu protein gp35, Protein HI1507 in Mu-like prophage FluMu region | | Authors: | Sari, N, He, Y, Doseeva, V, Surabian, K, Schwarz, F, Herzberg, O, Orban, J, Structure 2 Function Project (S2F) | | Deposit date: | 2007-02-12 | | Release date: | 2007-05-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of HI1506, a novel two-domain protein from Haemophilus influenzae.
Protein Sci., 16, 2007
|
|
2KT7
 
 | | Solution NMR structure of mucin-binding domain of protein lmo0835 from Listeria monocytogenes, Northeast Structural Genomics Consortium Target LmR64A | | Descriptor: | Putative peptidoglycan bound protein (LPXTG motif) | | Authors: | Eletsky, A, He, Y, Lee, D, Ciccosanti, C, Janjua, H, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-01-20 | | Release date: | 2010-02-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of mucin-binding domain of protein lmo0835 from Listeria monocytogenes
To be Published
|
|
6INN
 
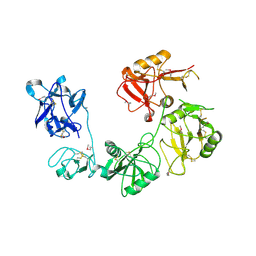 | |
6INV
 
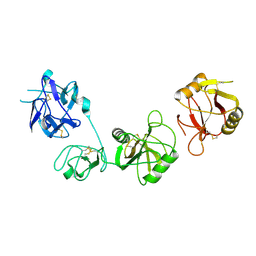 | |
6INO
 
 | |
6IOE
 
 | |
6INU
 
 | |
8V41
 
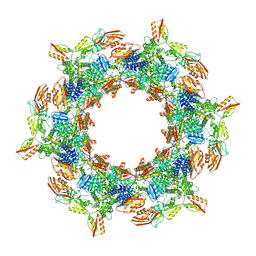 | | CryoEM Structure of Diffocin - postcontracted - Baseplate - transitional state | | Descriptor: | Sheath (CD1363), Sheath initiator (CD1370), TRI-1 (CD1372), ... | | Authors: | Cai, X.Y, He, Y, Zhou, Z.H. | | Deposit date: | 2023-11-28 | | Release date: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Atomic structures of a bacteriocin targeting Gram-positive bacteria.
Nat Commun, 15, 2024
|
|
8V3Y
 
 | |
8V3Z
 
 | |
8V3T
 
 | |
8V43
 
 | | CryoEM Structure of Diffocin - postcontracted - Baseplate - final state | | Descriptor: | Sheath (CD1363), Sheath initiator (CD1370), TRI-1 (CD1372), ... | | Authors: | Cai, X.Y, He, Y, Zhou, Z.H. | | Deposit date: | 2023-11-28 | | Release date: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Atomic structures of a bacteriocin targeting Gram-positive bacteria.
Nat Commun, 15, 2024
|
|
8V3W
 
 | | CryoEM Structure of Diffocin - precontracted - Baseplate - focused refinement on triplex region | | Descriptor: | Hub-Hydrolase (CD1368), Sheath (CD1363), Sheath initiator (CD1370), ... | | Authors: | Cai, X.Y, He, Y, Zhou, Z.H. | | Deposit date: | 2023-11-28 | | Release date: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Atomic structures of a bacteriocin targeting Gram-positive bacteria.
Nat Commun, 15, 2024
|
|
