1MT6
 
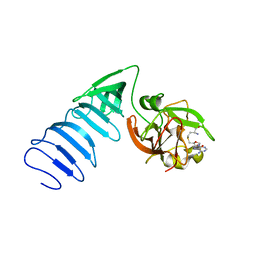 | | Structure of histone H3 K4-specific methyltransferase SET7/9 with AdoHcy | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SET9 | | Authors: | Jacobs, S.A, Harp, J.M, Devarakonda, S, Kim, Y, Rastinejad, F, Khorasanizadeh, S. | | Deposit date: | 2002-09-20 | | Release date: | 2002-11-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The active site of the SET domain is constructed on a knot
Nat.Struct.Biol., 9, 2002
|
|
4YP3
 
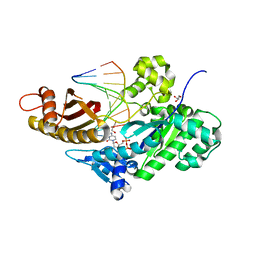 | | Mutant Human DNA Polymerase Eta Q38A/R61A Inserting dCTP Opposite an 8-Oxoguanine Lesion | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*T)-3'), ... | | Authors: | Su, Y, Patra, A, Harp, J.M, Egli, M, Guengerich, F.P. | | Deposit date: | 2015-03-12 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Roles of Residues Arg-61 and Gln-38 of Human DNA Polymerase eta in Bypass of Deoxyguanosine and 7,8-Dihydro-8-oxo-2'-deoxyguanosine.
J.Biol.Chem., 290, 2015
|
|
4YR0
 
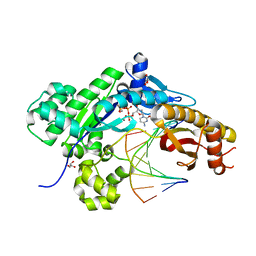 | | Mutant Human DNA Polymerase Eta R61M Inserting dCTP Opposite an 8-Oxoguanine Lesion | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*T)-3'), ... | | Authors: | Su, Y, Patra, A, Harp, J.M, Egli, M, Guengerich, F.P. | | Deposit date: | 2015-03-13 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Roles of Residues Arg-61 and Gln-38 of Human DNA Polymerase eta in Bypass of Deoxyguanosine and 7,8-Dihydro-8-oxo-2'-deoxyguanosine.
J.Biol.Chem., 290, 2015
|
|
4YR2
 
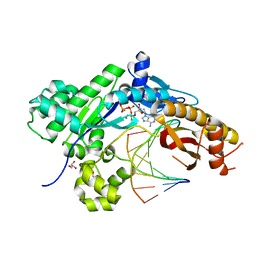 | | Mutant Human DNA Polymerase Eta R61M Inserting dATP Opposite an 8-Oxoguanine Lesion | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*T)-3'), ... | | Authors: | Su, Y, Patra, A, Harp, J.M, Egli, M, Guengerich, F.P. | | Deposit date: | 2015-03-14 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Roles of Residues Arg-61 and Gln-38 of Human DNA Polymerase eta in Bypass of Deoxyguanosine and 7,8-Dihydro-8-oxo-2'-deoxyguanosine.
J.Biol.Chem., 290, 2015
|
|
4YR3
 
 | | Mutant Human DNA Polymerase Eta R61M Inserting dCTP Opposite Template G | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*T)-3'), ... | | Authors: | Su, Y, Patra, A, Harp, J.M, Egli, M, Guengerich, F.P. | | Deposit date: | 2015-03-14 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Roles of Residues Arg-61 and Gln-38 of Human DNA Polymerase eta in Bypass of Deoxyguanosine and 7,8-Dihydro-8-oxo-2'-deoxyguanosine.
J.Biol.Chem., 290, 2015
|
|
7L5M
 
 | | Crystal Structure of the DiB-RM-split Protein | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Lipocalin family protein, SODIUM ION, ... | | Authors: | Bozhanova, N.G, Harp, J.M, Meiler, J. | | Deposit date: | 2020-12-22 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Computational redesign of a fluorogen activating protein with Rosetta.
Plos Comput.Biol., 17, 2021
|
|
7L5L
 
 | |
7L5K
 
 | | Crystal structure of the DiB-RM protein | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, ISOPROPYL ALCOHOL, Lipocalin family protein, ... | | Authors: | Bozhanova, N.G, Harp, J.M, Meiler, J. | | Deposit date: | 2020-12-22 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Computational redesign of a fluorogen activating protein with Rosetta.
Plos Comput.Biol., 17, 2021
|
|
3MFA
 
 | | Computationally designed endo-1,4-beta-xylanase | | Descriptor: | Endo-1,4-beta-xylanase, SULFATE ION | | Authors: | Morin, A, Harp, J.M. | | Deposit date: | 2010-04-01 | | Release date: | 2010-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Computational design of an endo-1,4-{beta}-xylanase ligand binding site.
Protein Eng.Des.Sel., 24, 2011
|
|
3MF9
 
 | | Computationally designed endo-1,4-beta-xylanase | | Descriptor: | Endo-1,4-beta-xylanase, SULFATE ION | | Authors: | Morin, A, Harp, J.M. | | Deposit date: | 2010-04-01 | | Release date: | 2010-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Computational design of an endo-1,4-{beta}-xylanase ligand binding site.
Protein Eng.Des.Sel., 24, 2011
|
|
3MFC
 
 | | Computationally designed end0-1,4-beta,xylanase | | Descriptor: | Endo-1,4-beta-xylanase, SULFATE ION | | Authors: | Morin, A, Harp, J.M. | | Deposit date: | 2010-04-01 | | Release date: | 2010-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Computational design of an endo-1,4-{beta}-xylanase ligand binding site.
Protein Eng.Des.Sel., 24, 2011
|
|
3MF6
 
 | | Computationally designed endo-1,4-beta-xylanase | | Descriptor: | Endo-1,4-beta-xylanase, SULFATE ION | | Authors: | Morin, A, Harp, J.M. | | Deposit date: | 2010-04-01 | | Release date: | 2010-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Computational design of an endo-1,4-{beta}-xylanase ligand binding site.
Protein Eng.Des.Sel., 24, 2011
|
|
3GX0
 
 | | Crystal Structure of GSH-dependent Disulfide bond Oxidoreductase | | Descriptor: | GST-like protein yfcG, OXIDIZED GLUTATHIONE DISULFIDE | | Authors: | Ladner, J.E, Harp, J.M, Wadington, M.C, Armstrong, R.N. | | Deposit date: | 2009-04-01 | | Release date: | 2009-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Analysis of the structure and function of YfcG from Escherichia coli reveals an efficient and unique disulfide bond reductase.
Biochemistry, 48, 2009
|
|
3NTB
 
 | | Structure of 6-methylthio naproxen analog bound to mCOX-2. | | Descriptor: | (2S)-2-[6-(methylsulfanyl)naphthalen-2-yl]propanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Duggan, K.C, Musee, J, Walters, M.J, Harp, J.M, Kiefer, J.R, Oates, J.A, Marnett, L.J. | | Deposit date: | 2010-07-03 | | Release date: | 2010-09-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Molecular basis for cyclooxygenase inhibition by the non-steroidal anti-inflammatory drug naproxen.
J.Biol.Chem., 285, 2010
|
|
3NT1
 
 | | High resolution structure of naproxen:COX-2 complex. | | Descriptor: | (2S)-2-(6-methoxynaphthalen-2-yl)propanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Duggan, K.C, Musee, J, Walters, M.J, Harp, J.M, Kiefer, J.R, Oates, J.A, Marnett, L.J. | | Deposit date: | 2010-07-02 | | Release date: | 2010-09-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Molecular basis for cyclooxygenase inhibition by the non-steroidal anti-inflammatory drug naproxen.
J.Biol.Chem., 285, 2010
|
|
3R3E
 
 | | The glutathione bound structure of YqjG, a glutathione transferase homolog from Escherichia coli K-12 | | Descriptor: | GLUTATHIONE, SULFATE ION, Uncharacterized protein yqjG | | Authors: | Branch, M.C, Cook, P.D, Harp, J.M, Armstrong, R.N. | | Deposit date: | 2011-03-15 | | Release date: | 2012-03-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.205 Å) | | Cite: | Crystal Structure Analysis of yqjG from Escherichia Coli K-12
To be Published
|
|
1R0O
 
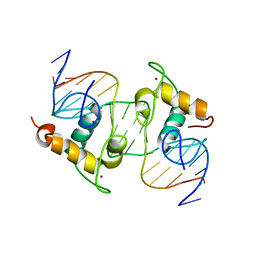 | | Crystal Structure of the Heterodimeric Ecdysone Receptor DNA-binding Complex | | Descriptor: | Ecdysone Response Element, Ecdysone receptor, Ultraspiracle protein, ... | | Authors: | Devarakonda, S, Harp, J.M, Kim, Y, Ozyhar, A, Rastinejad, F. | | Deposit date: | 2003-09-22 | | Release date: | 2003-10-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structure of the Heterodimeric Ecdysone Receptor DNA-binding Complex
Embo J., 22, 2003
|
|
1QCO
 
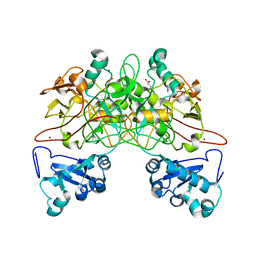 | | CRYSTAL STRUCTURE OF FUMARYLACETOACETATE HYDROLASE COMPLEXED WITH FUMARATE AND ACETOACETATE | | Descriptor: | ACETOACETIC ACID, CALCIUM ION, FUMARIC ACID, ... | | Authors: | Timm, D.E, Mueller, H.A, Bhanumoorthy, P, Harp, J.M, Bunick, G.J. | | Deposit date: | 1999-05-17 | | Release date: | 2000-06-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and mechanism of a carbon-carbon bond hydrolase.
Structure Fold.Des., 7, 1999
|
|
1R0N
 
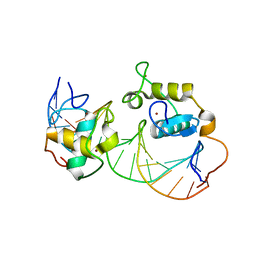 | | Crystal Structure of Heterodimeric Ecdsyone receptor DNA binding complex | | Descriptor: | Ecdsyone Response Element, Ecdysone Response Element, Ecdysone receptor, ... | | Authors: | Devarakonda, S, Harp, J.M, Kim, Y, Ozyhar, A, Rastinejad, F. | | Deposit date: | 2003-09-22 | | Release date: | 2003-10-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the heterodimeric Ecdysone Receptor DNA-binding complex
Embo J., 22, 2003
|
|
1OSV
 
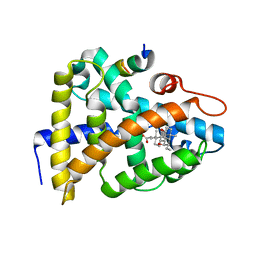 | | STRUCTURAL BASIS FOR BILE ACID BINDING AND ACTIVATION OF THE NUCLEAR RECEPTOR FXR | | Descriptor: | 6-ETHYL-CHENODEOXYCHOLIC ACID, Bile acid receptor, Nuclear receptor coactivator 2 | | Authors: | Mi, L.Z, Devarakonda, S, Harp, J.M, Han, Q, Pellicciari, R, Willson, T.M, Khorasanizadeh, S, Rastinejad, F. | | Deposit date: | 2003-03-20 | | Release date: | 2004-03-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Bile Acid Binding and Activation of the Nuclear Receptor FXR
Mol.Cell, 11, 2003
|
|
1QQJ
 
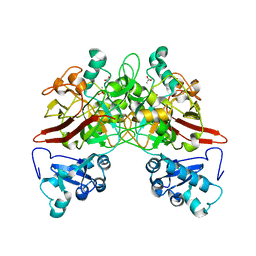 | | CRYSTAL STRUCTURE OF MOUSE FUMARYLACETOACETATE HYDROLASE REFINED AT 1.55 ANGSTROM RESOLUTION | | Descriptor: | ACETATE ION, CACODYLATE ION, CALCIUM ION, ... | | Authors: | Timm, D.E, Mueller, H.A, Bhanumoorthy, P, Harp, J.M, Bunick, G.J. | | Deposit date: | 1999-06-07 | | Release date: | 2000-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure and mechanism of a carbon-carbon bond hydrolase.
Structure Fold.Des., 7, 1999
|
|
1QCN
 
 | | CRYSTAL STRUCTURE OF FUMARYLACETOACETATE HYDROLASE | | Descriptor: | ACETATE ION, CALCIUM ION, FUMARYLACETOACETATE HYDROLASE, ... | | Authors: | Timm, D.E, Mueller, H.A, Bhanumoorthy, P, Harp, J.M, Bunick, G.J. | | Deposit date: | 1999-05-14 | | Release date: | 2000-06-07 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and mechanism of a carbon-carbon bond hydrolase.
Structure Fold.Des., 7, 1999
|
|
1OT7
 
 | | Structural Basis for 3-deoxy-CDCA Binding and Activation of FXR | | Descriptor: | 6-ETHYL-CHENODEOXYCHOLIC ACID, Bile Acid Receptor, ISO-URSODEOXYCHOLIC ACID, ... | | Authors: | Mi, L.Z, Devarakonda, S, Harp, J.M, Han, Q, Pellicciari, R, Willson, T.M, Khorasanizadeh, S, Rastinejad, F. | | Deposit date: | 2003-03-21 | | Release date: | 2004-03-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Basis for Bile Acid Binding and Activation of the Nuclear Receptor FXR
Mol.Cell, 11, 2003
|
|
2GLK
 
 | | High-resolution study of D-Xylose isomerase, 0.94A resolution. | | Descriptor: | GLYCEROL, MANGANESE (II) ION, Xylose isomerase | | Authors: | Katz, A.K, Carrell, H.L, Hanson, B.L, Harp, J.M, Glusker, J.P, Bunick, G.J. | | Deposit date: | 2006-04-05 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Locating active-site hydrogen atoms in D-xylose isomerase: Time-of-flight neutron diffraction.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
8UPT
 
 | | Candidatus Methanomethylophilus alvus tRNAPyl in A-site of ribosome | | Descriptor: | RNA (71-MER) | | Authors: | Krahn, N, Zhang, J, Melnikov, S.V, Tharp, J.M, Villa, A, Patel, A, Howard, R.J, Gabir, H, Patel, T.R, Stetefeld, J, Puglisi, J, Soll, D. | | Deposit date: | 2023-10-23 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | tRNA shape is an identity element for an archaeal pyrrolysyl-tRNA synthetase from the human gut.
Nucleic Acids Res., 52, 2024
|
|
