6IJF
 
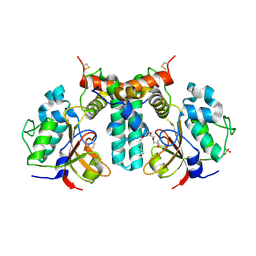 | | Crystal structure of the type VI effector-immunity complex (Tae4-Tai4) from Agrobacterium tumefaciens | | Descriptor: | PENTAETHYLENE GLYCOL, SULFATE ION, Tae4, ... | | Authors: | Fukuhara, S, Nakane, T, Yamashita, K, Ishii, R, Ishitani, R, Nureki, O. | | Deposit date: | 2018-10-09 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the Agrobacterium tumefaciens type VI effector-immunity complex.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6IJE
 
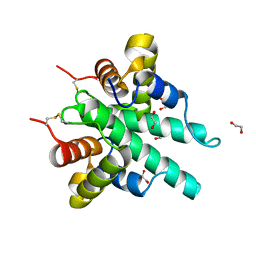 | | Crystal structure of the type VI amidase immunity (Tai4) from Agrobacterium tumefaciens | | Descriptor: | 1,2-ETHANEDIOL, Tai4 | | Authors: | Fukuhara, S, Nakane, T, Yamashita, K, Ishii, R, Ishitani, R, Nureki, O. | | Deposit date: | 2018-10-09 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of the Agrobacterium tumefaciens type VI effector-immunity complex.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
3R0Z
 
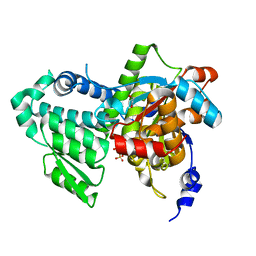 | | Crystal structure of apo D-serine deaminase from Salmonella typhimurium | | Descriptor: | 1,2-ETHANEDIOL, D-serine dehydratase, SULFATE ION | | Authors: | Bharath, S.R, Shveta, B, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2011-03-09 | | Release date: | 2011-06-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of open and closed forms of D-serine deaminase from Salmonella typhimurium - implications on substrate specificity and catalysis
Febs J., 2011
|
|
3QQD
 
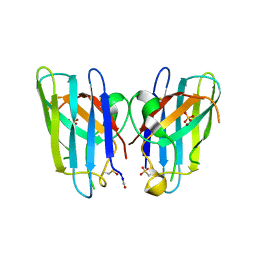 | | Human SOD1 H80R variant, P212121 crystal form | | Descriptor: | SULFATE ION, Superoxide dismutase [Cu-Zn], ZINC ION | | Authors: | Seetharaman, S.V, Winkler, D.D, Taylor, A.B, Cao, X, Whitson, L.J, Doucette, P.A, Valentine, J.S, Schirf, V, Demeler, B, Carroll, M.C, Culotta, V.C, Hart, P.J. | | Deposit date: | 2011-02-15 | | Release date: | 2011-03-09 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.653 Å) | | Cite: | Disrupted zinc-binding sites in structures of pathogenic SOD1 variants D124V and H80R.
Biochemistry, 49, 2010
|
|
3R0X
 
 | | Crystal structure of Selenomethionine incorporated apo D-serine deaminase from Salmonella tyhimurium | | Descriptor: | 1,2-ETHANEDIOL, D-serine dehydratase, SODIUM ION, ... | | Authors: | Bharath, S.R, Shveta, B, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2011-03-09 | | Release date: | 2011-06-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structures of open and closed forms of D-serine deaminase from Salmonella typhimurium - implications on substrate specificity and catalysis
Febs J., 2011
|
|
3HWO
 
 | |
3N35
 
 | | Erythrina corallodendron lectin mutant (Y106G) with N-Acetylgalactosamine | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, CALCIUM ION, Lectin, ... | | Authors: | Thamotharan, S, Karthikeyan, T, Kulkarni, K.A, Shetty, K.N, Surolia, A, Vijayan, M, Suguna, K. | | Deposit date: | 2010-05-19 | | Release date: | 2011-03-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Modification of the sugar specificity of a plant lectin: structural studies on a point mutant of Erythrina corallodendron lectin.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3N36
 
 | | Erythrina corallodendron lectin mutant (Y106G) in complex with Galactose | | Descriptor: | CALCIUM ION, Lectin, MANGANESE (II) ION, ... | | Authors: | Thamotharan, S, Karthikeyan, T, Kulkarni, K.A, Shetty, K.N, Surolia, A, Vijayan, M, Suguna, K. | | Deposit date: | 2010-05-19 | | Release date: | 2011-03-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Modification of the sugar specificity of a plant lectin: structural studies on a point mutant of Erythrina corallodendron lectin.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3N3H
 
 | | Erythrina corallodendron lectin mutant (Y106G) in complex with citrate | | Descriptor: | CALCIUM ION, CITRIC ACID, Lectin, ... | | Authors: | Thamotharan, S, Karthikeyan, T, Kulkarni, K.A, Shetty, K.N, Surolia, A, Vijayan, M, Suguna, K. | | Deposit date: | 2010-05-20 | | Release date: | 2011-03-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Modification of the sugar specificity of a plant lectin: structural studies on a point mutant of Erythrina corallodendron lectin.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
1V66
 
 | | Solution structure of human p53 binding domain of PIAS-1 | | Descriptor: | Protein inhibitor of activated STAT protein 1 | | Authors: | Okubo, S, Hara, F, Tsuchida, Y, Shimotakahara, S, Suzuki, S, Hatanaka, H, Yokoyama, S, Tanaka, H, Yasuda, H, Shindo, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-27 | | Release date: | 2004-12-07 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the N-terminal domain of SUMO ligase PIAS1 and its interaction with tumor suppressor p53 and A/T-rich DNA oligomers
J.Biol.Chem., 279, 2004
|
|
5XPT
 
 | |
5XPU
 
 | |
1IRH
 
 | | The Solution Structure of The Third Kunitz Domain of Tissue Factor Pathway Inhibitor | | Descriptor: | tissue factor pathway inhibitor | | Authors: | Mine, S, Yamazaki, T, Miyata, T, Hara, S, Kato, H. | | Deposit date: | 2001-10-02 | | Release date: | 2002-02-06 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural mechanism for heparin-binding of the third Kunitz domain of human tissue factor pathway inhibitor.
Biochemistry, 41, 2002
|
|
1IXU
 
 | | Solution structure of marinostatin, a protease inhibitor, containing two ester linkages | | Descriptor: | marinostatin | | Authors: | Kanaori, K, Kamei, K, Koyama, T, Yasui, T, Takano, R, Imada, C, Tajima, K, Hara, S. | | Deposit date: | 2002-07-04 | | Release date: | 2004-02-17 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of marinostatin, a natural ester-linked protein protease inhibitor
Biochemistry, 44, 2005
|
|
1KV4
 
 | | Solution structure of antibacterial peptide (Moricin) | | Descriptor: | moricin | | Authors: | Hemmi, H, Ishibashi, J, Hara, S, Yamakawa, M. | | Deposit date: | 2002-01-25 | | Release date: | 2002-05-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of moricin, an antibacterial peptide, isolated from the silkworm Bombyx mori.
FEBS Lett., 518, 2002
|
|
4UU9
 
 | | Crystal structure of the human c5a in complex with MEDI7814 a neutralising antibody | | Descriptor: | COMPLEMENT C5, MEDI7814, SULFATE ION | | Authors: | Colley, C, Sridharan, S, Dobson, C, Popovic, B, Debreczeni, J.E, Hargreaves, D, Edwards, B, Brennan, J, England, L, Fung, S, An Eghobamien, L, Sivars, U, Woods, R, Flavell, L, Renshaw, G.J, Wickson, K, Wilkinson, T, Davies, R, Bonnell, J, Warrener, P, Howes, R, Vaughan, T. | | Deposit date: | 2014-07-25 | | Release date: | 2015-08-12 | | Last modified: | 2019-02-27 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure and characterization of a high affinity C5a monoclonal antibody that blocks binding to C5aR1 and C5aR2 receptors.
MAbs, 10, 2018
|
|
4V95
 
 | | Crystal structure of YAEJ bound to the 70S ribosome | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | Authors: | Gagnon, M.G, Seetharaman, S.V, Bulkley, D.P, Steitz, T.A. | | Deposit date: | 2012-01-27 | | Release date: | 2014-07-09 | | Last modified: | 2018-07-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for the rescue of stalled ribosomes: structure of YaeJ bound to the ribosome.
Science, 335, 2012
|
|
1UHM
 
 | | Solution structure of the globular domain of linker histone homolog Hho1p from S. cerevisiae | | Descriptor: | Histone H1 | | Authors: | Ono, K, Kusano, O, Shimotakahara, S, Shimizu, M, Yamazaki, T, Shindo, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-05 | | Release date: | 2003-12-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The linker histone homolog Hho1p from Saccharomyces cerevisiae represents a winged helix-turn-helix fold as determined by NMR spectroscopy.
Nucleic Acids Res., 31, 2003
|
|
1A0F
 
 | | CRYSTAL STRUCTURE OF GLUTATHIONE S-TRANSFERASE FROM ESCHERICHIA COLI COMPLEXED WITH GLUTATHIONESULFONIC ACID | | Descriptor: | GLUTATHIONE S-TRANSFERASE, GLUTATHIONE SULFONIC ACID | | Authors: | Nishida, M, Harada, S, Noguchi, S, Inoue, H, Takahashi, K, Satow, Y. | | Deposit date: | 1997-11-29 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Three-dimensional structure of Escherichia coli glutathione S-transferase complexed with glutathione sulfonate: catalytic roles of Cys10 and His106.
J.Mol.Biol., 281, 1998
|
|
2E6F
 
 | | Crystal structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with oxonate | | Descriptor: | COBALT HEXAMMINE(III), Dihydroorotate dehydrogenase, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Inaoka, D.K, Shimizu, H, Sakamoto, K, Shiba, T, Kurisu, G, Nara, T, Aoki, T, Harada, S, Kita, K. | | Deposit date: | 2006-12-26 | | Release date: | 2008-01-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Crystal structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with oxonate
To be Published
|
|
2E68
 
 | | Crystal structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with dihydroorotate | | Descriptor: | (4S)-2,6-DIOXOHEXAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, COBALT HEXAMMINE(III), Dihydroorotate dehydrogenase, ... | | Authors: | Inaoka, D.K, Shimizu, H, Sakamoto, K, Shiba, T, Kurisu, G, Nara, T, Aoki, T, Harada, S, Kita, K. | | Deposit date: | 2006-12-26 | | Release date: | 2008-01-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Crystal structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with dihydroorotate
To be Published
|
|
4UV4
 
 | | Crystal structure of anti-FPR Fpro0165 Fab fragment | | Descriptor: | FPRO0165 FAB | | Authors: | Douthwaite, J.A, Sridharan, S, Huntington, C, Marwood, R, Hammersley, J, Hakulinen, J.K, Ek, M, Sjogren, T, Rider, D, Privezentzev, C, Seaman, J.C, Cariuk, P, Knights, V, Young, J, Wilkinson, T, Sleeman, M, Finch, D.K, Lowe, D.C, Vaughan, T.J. | | Deposit date: | 2014-08-04 | | Release date: | 2014-12-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Affinity Maturation of a Novel Antagonistic Human Monoclonal Antibody with a Long Vh Cdr3 Targeting the Class a Gpcr Formyl-Peptide Receptor 1.
Mabs, 7, 2015
|
|
8SEI
 
 | | SF Tau from Down Syndrome | | Descriptor: | Microtubule-associated protein tau | | Authors: | Hoq, M.R, Bharath, S.R, Jiang, W, Vago, F.S, Bharath, S.R. | | Deposit date: | 2023-04-10 | | Release date: | 2024-04-03 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures of amyloid-beta and tau filaments in Down syndrome.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8F9K
 
 | |
8SEL
 
 | |
