8IMG
 
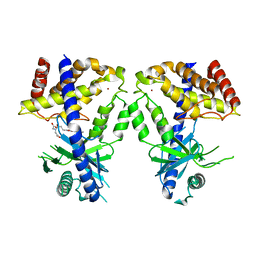 | | Human cGAS catalytic domain bound with C20 | | Descriptor: | 2-[2-hydroxy-2-oxoethyl-[3-(7-methoxy-4-methyl-2-oxidanylidene-chromen-3-yl)propanoyl]amino]ethanoic acid, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Zhao, W.F, Li, J.M, Xu, Y.C. | | Deposit date: | 2023-03-06 | | Release date: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of novel cGAS inhibitors based on natural flavonoids.
Bioorg.Chem., 140, 2023
|
|
4M70
 
 | |
7FEX
 
 | |
4AEZ
 
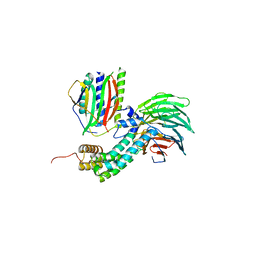 | | Crystal Structure of Mitotic Checkpoint Complex | | Descriptor: | MITOTIC SPINDLE CHECKPOINT COMPONENT MAD2, MITOTIC SPINDLE CHECKPOINT COMPONENT MAD3, WD REPEAT-CONTAINING PROTEIN SLP1 | | Authors: | Kulkarni, K.A, Chao, W.C.H, Zhang, Z, Barford, D. | | Deposit date: | 2012-01-13 | | Release date: | 2012-03-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Mitotic Checkpoint Complex
Nature, 484, 2012
|
|
6U5G
 
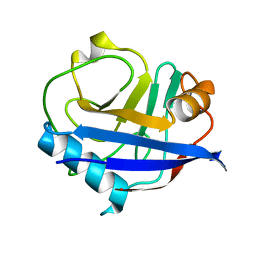 | | MicroED structure of a FIB-milled CypA Crystal | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A | | Authors: | Wolff, A.M, Martynowycz, M.W, Zhao, W, Gonen, T, Fraser, J.S, Thompson, M.C. | | Deposit date: | 2019-08-27 | | Release date: | 2020-01-29 | | Last modified: | 2023-10-11 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.5 Å) | | Cite: | Comparing serial X-ray crystallography and microcrystal electron diffraction (MicroED) as methods for routine structure determination from small macromolecular crystals
Iucrj, 7, 2020
|
|
4UX5
 
 | | Structure of DNA complex of PCG2 | | Descriptor: | 5'-D(*CP*AP*AP*TP*GP*AP*CP*GP*CP*GP*TP*AP*AP*GP)-3', 5'-D(*CP*TP*TP*AP*CP*GP*CP*GP*TP*CP*AP*TP*TP*GP)-3', TRANSCRIPTION FACTOR MBP1 | | Authors: | Liu, J, Huang, J, Zhao, Y, Liu, H, Wang, D, Yang, J, Zhao, W, Taylor, I.A, Peng, Y. | | Deposit date: | 2014-08-19 | | Release date: | 2015-01-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of DNA Recognition by Pcg2 Reveals a Novel DNA Binding Mode for Winged Helix-Turn-Helix Domains.
Nucleic Acids Res., 43, 2015
|
|
8Z1R
 
 | | isocitrate lyase MoMcl1 | | Descriptor: | 1,2-ETHANEDIOL, Isocitrate lyase, MAGNESIUM ION | | Authors: | Liu, X.H, Zhao, W.H. | | Deposit date: | 2024-04-11 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | High-resolution crystal structure of the MoMcl1 protein
To Be Published
|
|
8Z28
 
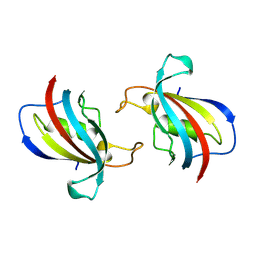 | | FK506-binding protein 1B | | Descriptor: | peptidylprolyl isomerase | | Authors: | Liu, X.H, Zhao, W.H. | | Deposit date: | 2024-04-12 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | FK506-binding protein 1B
To Be Published
|
|
8Z38
 
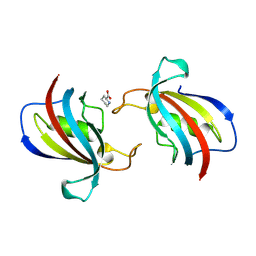 | | FK506 binding protein 1B (including FK506) | | Descriptor: | 6-CARBOXYPIPERIDINE, peptidylprolyl isomerase | | Authors: | Liu, X.H, Zhao, W.H. | | Deposit date: | 2024-04-14 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | High-resolution cocrystallized crystal structures of FK506 and FK506-binding proteins
To Be Published
|
|
6M2N
 
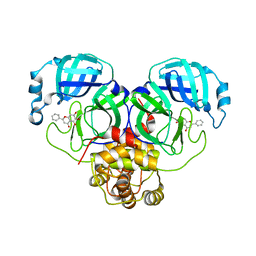 | | SARS-CoV-2 3CL protease (3CL pro) in complex with a novel inhibitor | | Descriptor: | 3C-like proteinase, 5,6,7-trihydroxy-2-phenyl-4H-chromen-4-one | | Authors: | Su, H.X, Zhao, W.F, Li, M.J, Xie, H, Xu, Y.C. | | Deposit date: | 2020-02-28 | | Release date: | 2020-04-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Anti-SARS-CoV-2 activities in vitro of Shuanghuanglian preparations and bioactive ingredients.
Acta Pharmacol.Sin., 41, 2020
|
|
6M2Q
 
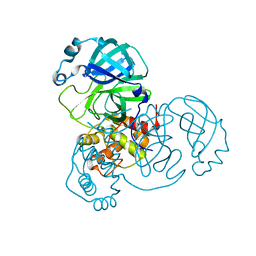 | | SARS-CoV-2 3CL protease (3CL pro) apo structure (space group C21) | | Descriptor: | 3C-like proteinase | | Authors: | Su, H.X, Zhao, W.F, Li, M.J, Xie, H, Xu, Y.C. | | Deposit date: | 2020-02-28 | | Release date: | 2020-04-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Anti-SARS-CoV-2 activities in vitro of Shuanghuanglian preparations and bioactive ingredients.
Acta Pharmacol.Sin., 41, 2020
|
|
5H1B
 
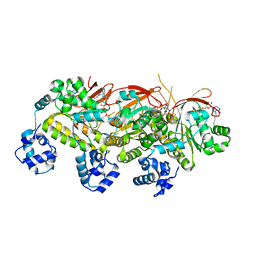 | | Human RAD51 presynaptic complex | | Descriptor: | DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), DNA repair protein RAD51 homolog 1, MAGNESIUM ION, ... | | Authors: | Xu, J, Zhao, L, Xu, Y, Zhao, W, Sung, P, Wang, H.W. | | Deposit date: | 2016-10-08 | | Release date: | 2016-12-21 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM structures of human RAD51 recombinase filaments during catalysis of DNA-strand exchange
Nat. Struct. Mol. Biol., 24, 2017
|
|
5H1C
 
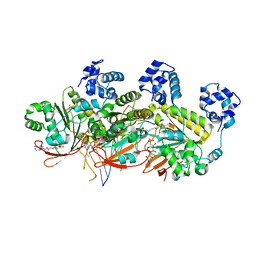 | | Human RAD51 post-synaptic complexes | | Descriptor: | DNA (5'-D(P*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), DNA repair protein RAD51 homolog 1, ... | | Authors: | Xu, J, Zhao, L, Xu, Y, Zhao, W, Sung, P, Wang, H.W. | | Deposit date: | 2016-10-08 | | Release date: | 2016-12-21 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM structures of human RAD51 recombinase filaments during catalysis of DNA-strand exchange
Nat. Struct. Mol. Biol., 24, 2017
|
|
6N4U
 
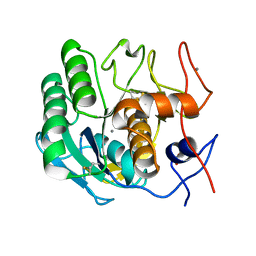 | | MicroED structure of Proteinase K at 2.75A resolution from a single milled crystal. | | Descriptor: | CALCIUM ION, Proteinase K, SULFATE ION | | Authors: | Martynowycz, M.W, Zhao, W, Hattne, J, Jensen, G.J, Gonen, T. | | Deposit date: | 2018-11-20 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.75 Å) | | Cite: | Collection of Continuous Rotation MicroED Data from Ion Beam-Milled Crystals of Any Size.
Structure, 27, 2019
|
|
5G06
 
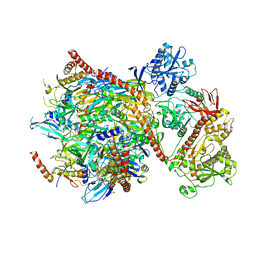 | | Cryo-EM structure of yeast cytoplasmic exosome | | Descriptor: | EXOSOME COMPLEX COMPONENT CSL4, EXOSOME COMPLEX COMPONENT MTR3, EXOSOME COMPLEX COMPONENT RRP4, ... | | Authors: | Liu, J.J, Niu, C.Y, Wu, Y, Tan, D, Wang, Y, Ye, M.D, Liu, Y, Zhao, W.W, Zhou, K, Liu, Q.S, Dai, J.B, Yang, X.R, Dong, M.Q, Huang, N, Wang, H.W. | | Deposit date: | 2016-03-17 | | Release date: | 2016-06-15 | | Last modified: | 2017-08-02 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryoem Structure of Yeast Cytoplasmic Exosome Complex.
Cell Res., 26, 2016
|
|
8WCP
 
 | |
6PKQ
 
 | | MicroED structure of proteinase K from a platinum-coated, polished, single lamella at 1.85A resolution (#11) | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Zhao, W, Hattne, J, Jensen, G.J, Gonen, T. | | Deposit date: | 2019-06-29 | | Release date: | 2019-09-04 | | Last modified: | 2019-12-18 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.85 Å) | | Cite: | Qualitative Analyses of Polishing and Precoating FIB Milled Crystals for MicroED.
Structure, 27, 2019
|
|
6PKM
 
 | | MicroED structure of proteinase K from an uncoated, single lamella at 2.17A resolution (#8) | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Zhao, W, Hattne, J, Jensen, G.J, Gonen, T. | | Deposit date: | 2019-06-29 | | Release date: | 2019-09-04 | | Last modified: | 2019-12-18 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.17 Å) | | Cite: | Qualitative Analyses of Polishing and Precoating FIB Milled Crystals for MicroED.
Structure, 27, 2019
|
|
6PKS
 
 | | MicroED structure of proteinase K from low-dose merged lamellae that were not pre-coated with platinum 2.16A resolution (LD) | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Zhao, W, Hattne, J, Jensen, G.J, Gonen, T. | | Deposit date: | 2019-06-29 | | Release date: | 2019-09-04 | | Last modified: | 2024-10-09 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.16 Å) | | Cite: | Qualitative Analyses of Polishing and Precoating FIB Milled Crystals for MicroED.
Structure, 27, 2019
|
|
6PKL
 
 | | MicroED structure of proteinase K from an uncoated, single lamella at 2.59A resolution (#7) | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Zhao, W, Hattne, J, Jensen, G.J, Gonen, T. | | Deposit date: | 2019-06-29 | | Release date: | 2019-09-04 | | Last modified: | 2019-12-18 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.59 Å) | | Cite: | Qualitative Analyses of Polishing and Precoating FIB Milled Crystals for MicroED.
Structure, 27, 2019
|
|
6PKT
 
 | | MicroED structure of proteinase K from merging low-dose, platinum pre-coated lamellae at 1.85A resolution (LDPT) | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Zhao, W, Hattne, J, Jensen, G.J, Gonen, T. | | Deposit date: | 2019-06-29 | | Release date: | 2019-09-04 | | Last modified: | 2024-10-23 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.85 Å) | | Cite: | Qualitative Analyses of Polishing and Precoating FIB Milled Crystals for MicroED.
Structure, 27, 2019
|
|
6PKJ
 
 | | MicroED structure of proteinase K from an uncoated, single lamella at 2.17A resolution (#2) | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Zhao, W, Hattne, J, Jensen, G.J, Gonen, T. | | Deposit date: | 2019-06-29 | | Release date: | 2019-09-04 | | Last modified: | 2019-12-18 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.176 Å) | | Cite: | Qualitative Analyses of Polishing and Precoating FIB Milled Crystals for MicroED.
Structure, 27, 2019
|
|
6PKO
 
 | | MicroED structure of proteinase K from a platinum coated, unpolished, single lamella at 2.07A resolution (#12) | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Zhao, W, Hattne, J, Jensen, G.J, Gonen, T. | | Deposit date: | 2019-06-29 | | Release date: | 2019-09-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.07 Å) | | Cite: | Qualitative Analyses of Polishing and Precoating FIB Milled Crystals for MicroED.
Structure, 27, 2019
|
|
6PKN
 
 | | MicroED structure of proteinase K from an unpolished, platinum-coated, single lamella at 2.08A resolution (#9) | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Zhao, W, Hattne, J, Jensen, G.J, Gonen, T. | | Deposit date: | 2019-06-29 | | Release date: | 2019-09-04 | | Last modified: | 2019-12-18 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.08 Å) | | Cite: | Qualitative Analyses of Polishing and Precoating FIB Milled Crystals for MicroED.
Structure, 27, 2019
|
|
6PKR
 
 | | MicroED structure of proteinase K from a platinum-coated, polished, single lamella at 1.79A resolution (#13) | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Zhao, W, Hattne, J, Jensen, G.J, Gonen, T. | | Deposit date: | 2019-06-29 | | Release date: | 2019-09-04 | | Last modified: | 2024-10-09 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.79 Å) | | Cite: | Qualitative Analyses of Polishing and Precoating FIB Milled Crystals for MicroED.
Structure, 27, 2019
|
|
