5ZEY
 
 | | M. smegmatis Trans-translation state 70S ribosome | | Descriptor: | A-tRNAfMet, SsrA-binding protein, tmRNA | | Authors: | Mishra, S, Ahmed, T, Tyagi, A, Shi, J, Bhushan, S. | | Deposit date: | 2018-02-28 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (12.5 Å) | | Cite: | Structures of Mycobacterium smegmatis 70S ribosomes in complex with HPF, tmRNA, and P-tRNA.
Sci Rep, 8, 2018
|
|
5I3D
 
 | | Sulfolobus solfataricus beta-glycosidase - E387Y mutant | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, ACETATE ION, Beta-galactosidase | | Authors: | Iglesias-Fernandez, J, Hancock, S.M, Lee, S.S, McAuley, K.E, Fordham-Skelton, A, Rovira, C, Davis, B.D. | | Deposit date: | 2016-02-10 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | A front-face 'SNi synthase' engineered from a retaining 'double-SN2' hydrolase.
Nat. Chem. Biol., 13, 2017
|
|
1LMR
 
 | | Solution of ADO1, a Toxin from the Assassin Bugs Agriosphodrus dohrni that Blocks the Voltage Sensitive Calcium Channel L-type | | Descriptor: | TOXIN ADO1 | | Authors: | Bernard, C, Corzo, G, Adachi-Akahane, S, Foures, G, Kanemaru, K, Furukawa, Y, Nakajima, T, Darbon, H. | | Deposit date: | 2002-05-02 | | Release date: | 2003-08-19 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of ADO1, a toxin extracted from the saliva of the assassin bug, Agriosphodrus dohrni
Proteins: STRUCT.,FUNCT.,GENET., 54, 2004
|
|
3MQT
 
 | |
4OHD
 
 | | LEOPARD Syndrome-Associated SHP2/A461T mutant | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Yu, Z.H, Zhang, R.Y, Walls, C.D, Chen, L, Zhang, S, Wu, L, Wang, L, Liu, S, Zhang, Z.Y. | | Deposit date: | 2014-01-17 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis of gain-of-function LEOPARD syndrome-associated SHP2 mutations.
Biochemistry, 53, 2014
|
|
3M0F
 
 | |
5DO0
 
 | | The structure of PKMT1 from Rickettsia prowazekii | | Descriptor: | protein lysine methyltransferase 1 | | Authors: | Noinaj, N, Abeykoon, A, He, Y, Yang, D.C, Buchanan, S.K. | | Deposit date: | 2015-09-10 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Insights into Substrate Recognition and Catalysis in Outer Membrane Protein B (OmpB) by Protein-lysine Methyltransferases from Rickettsia.
J.Biol.Chem., 291, 2016
|
|
3M8N
 
 | |
7CKR
 
 | | Cryo-EM structure of the human MCT1/Basigin-2 complex in the presence of anti-cancer drug candidate BAY-8002 in the outward-open conformation. | | Descriptor: | 2-[[2-chloranyl-5-(phenylsulfonyl)phenyl]carbonylamino]benzoic acid, Basigin, Monocarboxylate transporter 1 | | Authors: | Wang, N, Jiang, X, Zhang, S, Zhu, A, Yuan, Y, Lei, J, Yan, C. | | Deposit date: | 2020-07-18 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of human monocarboxylate transporter 1 inhibition by anti-cancer drug candidates.
Cell, 184, 2021
|
|
5VOB
 
 | |
4FB5
 
 | |
5E3M
 
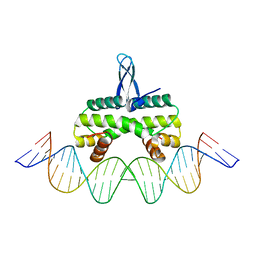 | | Crystal structure of Fis bound to 27bp DNA F35 (AAATTAGTTTGAATCTCGAGCTAATTT) | | Descriptor: | DNA (27-MER), DNA-binding protein Fis | | Authors: | Stella, S, Hancock, S.P, Cascio, D, Johnson, R.C. | | Deposit date: | 2015-10-03 | | Release date: | 2016-03-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.886 Å) | | Cite: | DNA Sequence Determinants Controlling Affinity, Stability and Shape of DNA Complexes Bound by the Nucleoid Protein Fis.
Plos One, 11, 2016
|
|
5VOC
 
 | |
4NJV
 
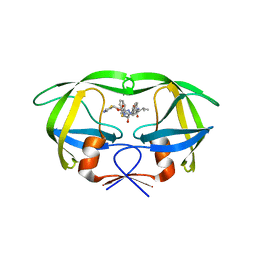 | | Crystal structure of multidrug-resistant clinical isolate A02 HIV-1 protease in complex with ritonavir | | Descriptor: | Protease, RITONAVIR | | Authors: | Yedidi, R.S, Garimella, H, Chang, S.B, Kaufman, J.D, Das, D, Wingfield, P.T, Mitsuya, H. | | Deposit date: | 2013-11-11 | | Release date: | 2014-04-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Conserved Hydrogen-Bonding Network of P2 bis-Tetrahydrofuran-Containing HIV-1 Protease Inhibitors (PIs) with a Protease Active-Site Amino Acid Backbone Aids in Their Activity against PI-Resistant HIV.
Antimicrob.Agents Chemother., 58, 2014
|
|
1XG7
 
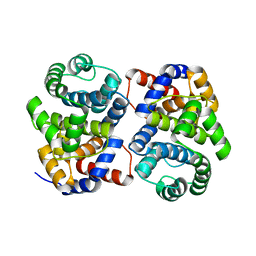 | | Conserved hypothetical protein Pfu-877259-001 from Pyrococcus furiosus | | Descriptor: | hypothetical protein | | Authors: | Chang, J, Zhao, M, Horanyi, P, Xu, H, Yang, H, Liu, Z.-J, Chen, L, Zhou, W, Habel, J, Tempel, W, Lee, D, Lin, D, Chang, S.-H, Eneh, J.C, Hopkins, R.C, Jenney Jr, F.E, Lee, H.-S, Li, T, Poole II, F.L, Shah, C, Sugar, F.J, Chen, C.-Y, Arendall III, W.B, Richardson, J.S, Richardson, D.C, Rose, J.P, Adams, M.W.W, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-09-16 | | Release date: | 2004-11-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Conserved hypothetical protein Pfu-877259-001 from Pyrococcus furiosus
To be published
|
|
3S1S
 
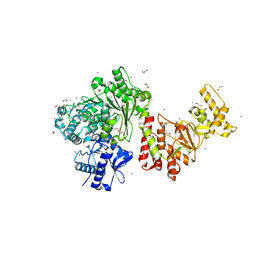 | | Characterization and crystal structure of the type IIG restriction endonuclease BpuSI | | Descriptor: | 1,2-ETHANEDIOL, IODIDE ION, MANGANESE (II) ION, ... | | Authors: | Shen, B.W, Xu, D, Chan, S.-H, Zheng, Y, Zhu, Y, Xu, S.-Y, Stoddard, B.L. | | Deposit date: | 2011-05-16 | | Release date: | 2011-07-13 | | Last modified: | 2011-10-19 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Characterization and crystal structure of the type IIG restriction endonuclease RM.BpuSI.
Nucleic Acids Res., 39, 2011
|
|
3KD9
 
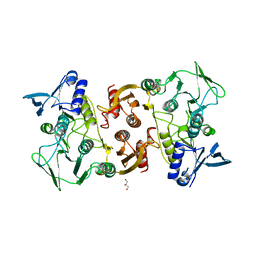 | |
4E3Z
 
 | | Crystal Structure of a oxidoreductase from Rhizobium etli CFN 42 | | Descriptor: | Putative oxidoreductase protein | | Authors: | Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-03-11 | | Release date: | 2012-03-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of a oxidoreductase from Rhizobium etli CFN 42
To be Published
|
|
5WAM
 
 | | Structure of BamE from Neisseria gonorrhoeae | | Descriptor: | Outer membrane protein assembly factor BamE, ZINC ION | | Authors: | Korotkov, K.V, Buchanan, S.K, Noinaj, N. | | Deposit date: | 2017-06-26 | | Release date: | 2017-12-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural and functional insights into the role of BamD and BamE within the beta-barrel assembly machinery in Neisseria gonorrhoeae.
J. Biol. Chem., 293, 2018
|
|
5WAQ
 
 | | Structure of BamD from Neisseria gonorrhoeae | | Descriptor: | Outer membrane protein assembly factor BamD | | Authors: | Korotkov, K.V, Buchanan, S.K, Noinaj, N. | | Deposit date: | 2017-06-26 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structural and functional insights into the role of BamD and BamE within the beta-barrel assembly machinery in Neisseria gonorrhoeae.
J. Biol. Chem., 293, 2018
|
|
2OOF
 
 | |
6JK3
 
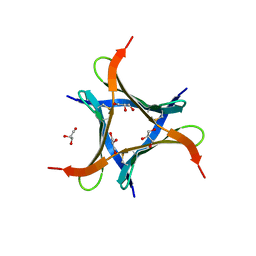 | | Crystal structure of a mini fungal lectin, PhoSL in complex with core-fucosylated chitobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Lectin | | Authors: | Lou, Y.C, Chou, C.C, Yeh, H.H, Chien, C.Y, Sushant, S, Chen, C, Hsu, C.H. | | Deposit date: | 2019-02-27 | | Release date: | 2020-03-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Structural insights into the role of N-terminal integrity in PhoSL for core-fucosylated N-glycan recognition.
Int.J.Biol.Macromol., 255, 2023
|
|
4EPA
 
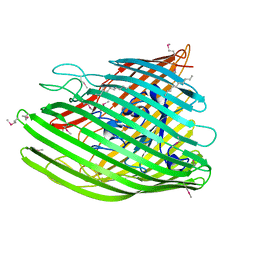 | |
1BF4
 
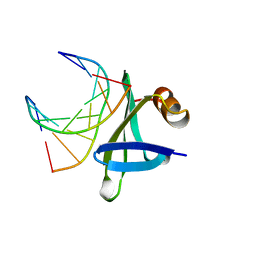 | | CHROMOSOMAL DNA-BINDING PROTEIN SSO7D/D(GCGAACGC) COMPLEX | | Descriptor: | DNA (5'-D(*GP*CP*GP*AP*AP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*TP*5IUP*CP*GP*C)-3'), PROTEIN (CHROMOSOMAL PROTEIN SSO7D) | | Authors: | Su, S, Gao, Y.-G, Robinson, H, Padmanabhan, S, Lim, L, Shriver, J.W, Wang, A.H.-J. | | Deposit date: | 1998-05-27 | | Release date: | 1999-11-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of the hyperthermophile chromosomal protein Sso7d bound to DNA.
Nat.Struct.Biol., 5, 1998
|
|
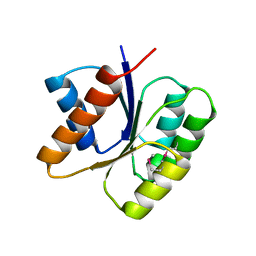
3KHT
 
 | |
