2EPE
 
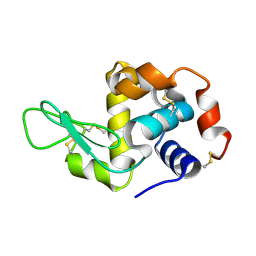 | | Crystal structure analysis of Hen egg white lysozyme grown by capillary method | | Descriptor: | Lysozyme C | | Authors: | Naresh, M.D, Subramanian, V, Jaimohan, S.M, Rajaram, A, Arumugam, V, Usha, R, Mandal, A.B. | | Deposit date: | 2007-03-29 | | Release date: | 2007-04-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure analysis of Hen egg white lysozyme grown by capillary method
To be published
|
|
1U4J
 
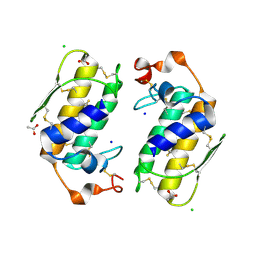 | | Crystal structure of a carbohydrate induced dimer of group I phospholipase A2 from Bungarus caeruleus at 2.1 A resolution | | Descriptor: | ACETIC ACID, CHLORIDE ION, SODIUM ION, ... | | Authors: | Singh, G, Gourinath, S, Sharma, S, Bhanumathi, S, Betzel, C, Srinivasan, A, Singh, T.P. | | Deposit date: | 2004-07-26 | | Release date: | 2004-08-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Crystal structure of a carbohydrate induced homodimer of phospholipase A(2) from Bungarus caeruleus at 2.1A resolution
J.Struct.Biol., 149, 2005
|
|
8E4Y
 
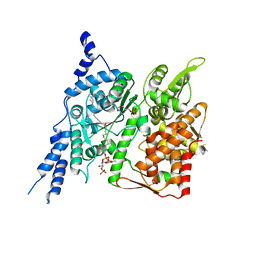 | | Cryo-EM structure of human glycerol-3-phosphate acyltransferase 1 (GPAT1) in complex with 2-oxohexadecyl-CoA | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl hexadecanoate, Glycerol-3-phosphate acyltransferase 1, mitochondrial, ... | | Authors: | Johnson, Z.L, Wasilko, D.J, Ammirati, M, Chang, J.S, Han, S, Wu, H. | | Deposit date: | 2022-08-19 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of the acyl-transfer mechanism of human GPAT1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
3FNR
 
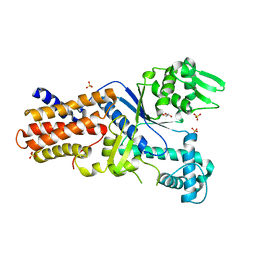 | | CRYSTAL STRUCTURE OF PUTATIVE ARGINYL T-RNA SYNTHETASE FROM Campylobacter jejuni; | | Descriptor: | Arginyl-tRNA synthetase, GLYCEROL, SULFATE ION | | Authors: | Patskovsky, Y, Ramagopal, U, Toro, R, Gilmore, M, Chang, S, Groshong, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-12-26 | | Release date: | 2009-01-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | CRYSTAL STRUCTURE OF A PUTATIVE ARGINYL T-RNA SYNTHETASE FROM Campylobacter jejuni
To be Published
|
|
1G9D
 
 | |
8E50
 
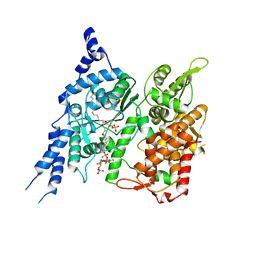 | | Cryo-EM structure of human glycerol-3-phosphate acyltransferase 1 (GPAT1) in complex with CoA and palmitoyl-LPA | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl hexadecanoate, COENZYME A, Glycerol-3-phosphate acyltransferase 1, ... | | Authors: | Wasilko, D.J, Johnson, Z.L, Ammirati, M, Chang, J.S, Han, S, Wu, H. | | Deposit date: | 2022-08-19 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Structural basis of the acyl-transfer mechanism of human GPAT1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8ASZ
 
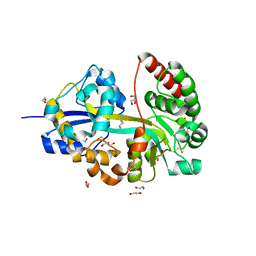 | |
1G9C
 
 | |
1G9B
 
 | |
3GHY
 
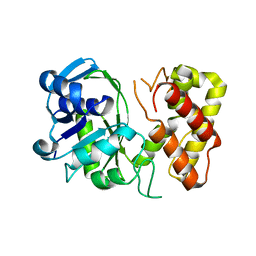 | | Crystal structure of a putative ketopantoate reductase from Ralstonia solanacearum MolK2 | | Descriptor: | Ketopantoate reductase protein | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Morano, C, Freeman, J, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-04 | | Release date: | 2009-03-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a putative ketopantoate reductase from Ralstonia solanacearum MolK2
To be Published
|
|
1G9A
 
 | |
3G0O
 
 | |
3GFG
 
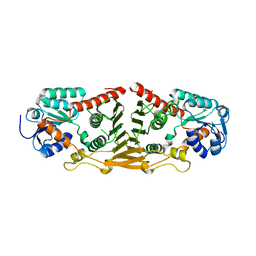 | | Structure of putative oxidoreductase yvaA from Bacillus subtilis in triclinic form | | Descriptor: | Uncharacterized oxidoreductase yvaA | | Authors: | Ramagopal, U.A, Toro, R, Gilmore, M, Chang, S, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-02-26 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure of putative oxidoreductase yvaA from Bacillus subtilis in triclinic form.
To be published
|
|
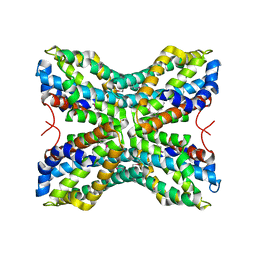
1TYH
 
 | |
1TY8
 
 | | Crystal structure of yeast ymx7, an ADP-ribose-1''-monophosphatase, complexed with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, Hypothetical 32.1 kDa protein in ADH3-RCA1 intergenic region, ... | | Authors: | Kumaran, D, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-07-07 | | Release date: | 2004-11-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | CRYSTAL STRUCTURE OF YEAST HYPOTHETICAL PROTEIN YMX7
To be Published
|
|
3FND
 
 | |
3TBL
 
 | | Structure of Mono-ubiquitinated PCNA: Implications for DNA Polymerase Switching and Okazaki Fragment Maturation | | Descriptor: | Proliferating cell nuclear antigen, Ubiquitin | | Authors: | Zhang, Z, Lee, M, Lee, E, Zhang, S. | | Deposit date: | 2011-08-07 | | Release date: | 2012-05-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.903 Å) | | Cite: | Structure of monoubiquitinated PCNA: Implications for DNA polymerase switching and Okazaki fragment maturation.
Cell Cycle, 11, 2012
|
|
6LMK
 
 | | Cryo-EM structure of the human glucagon receptor in complex with Gs | | Descriptor: | Glucagon, Glucagon receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Qiao, A, Han, S, Tai, L, Sun, F, Zhao, Q, Wu, B. | | Deposit date: | 2019-12-26 | | Release date: | 2020-04-01 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of Gsand Girecognition by the human glucagon receptor.
Science, 367, 2020
|
|
3G13
 
 | |
2PWT
 
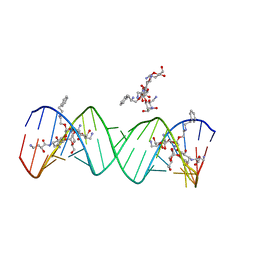 | | Crystal structure of the bacterial ribosomal decoding site complexed with aminoglycoside containing the L-HABA group | | Descriptor: | 22-mer of the ribosomal decoding site, DOUBLY FUNCTIONALIZED PAROMOMYCIN PM-II-162 | | Authors: | Kondo, J, Pachamuthu, K, Francois, B, Szychowski, J, Hanessian, S, Westhof, E. | | Deposit date: | 2007-05-13 | | Release date: | 2007-09-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Bacterial Ribosomal Decoding Site Complexed with a Synthetic Doubly Functionalized Paromomycin Derivative: a New Specific Binding Mode to an A-Minor Motif Enhances in vitro Antibacterial Activity
Chemmedchem, 2, 2007
|
|
6NZU
 
 | | Structure of the human frataxin-bound iron-sulfur cluster assembly complex | | Descriptor: | Acyl carrier protein, Cysteine desulfurase, mitochondrial, ... | | Authors: | Fox, N.G, Yu, X, Xidong, F, Alain, M, Joseph, N, Claire, S.D, Christine, B, Han, S, Yue, W.W. | | Deposit date: | 2019-02-14 | | Release date: | 2019-05-22 | | Last modified: | 2019-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the human frataxin-bound iron-sulfur cluster assembly complex provides insight into its activation mechanism.
Nat Commun, 10, 2019
|
|
1GHD
 
 | | Crystal structure of the glutaryl-7-aminocephalosporanic acid acylase by mad phasing | | Descriptor: | GLUTARYL-7-AMINOCEPHALOSPORANIC ACID ACYLASE | | Authors: | Ding, Y, Jiang, W, Mao, X, He, H, Zhang, S, Tang, H, Bartlam, M, Ye, S, Jiang, F, Liu, Y, Zhao, G, Rao, Z. | | Deposit date: | 2000-12-07 | | Release date: | 2003-07-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Affinity alkylation of the Trp-B4 residue of the beta -subunit of the glutaryl 7-aminocephalosporanic acid acylase of Pseudomonas sp. 130.
J.Biol.Chem., 277, 2002
|
|
6M2L
 
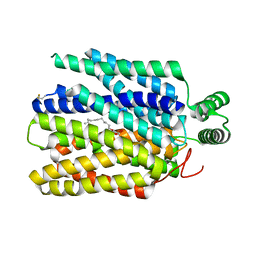 | | Crystal structure of Plasmodium falciparum hexose transporter PfHT1 bound with C3361 | | Descriptor: | (2S,3R,4S,5R,6R)-6-(hydroxymethyl)-4-undec-10-enoxy-oxane-2,3,5-triol, Hexose transporter 1 | | Authors: | Jiang, X, Yuan, Y.Y, Zhang, S, Wang, N, Yan, C.Y, Yan, N. | | Deposit date: | 2020-02-27 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural Basis for Blocking Sugar Uptake into the Malaria Parasite Plasmodium falciparum.
Cell, 183, 2020
|
|
6MP6
 
 | | Cryo-EM structure of the human neutral amino acid transporter ASCT2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Neutral amino acid transporter B(0) | | Authors: | Yu, X, Han, S. | | Deposit date: | 2018-10-05 | | Release date: | 2019-11-06 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Cryo-EM structures of the human glutamine transporter SLC1A5 (ASCT2) in the outward-facing conformation.
Elife, 8, 2019
|
|
6MPB
 
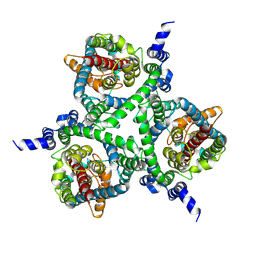 | | Cryo-EM structure of the human neutral amino acid transporter ASCT2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMINE, Neutral amino acid transporter B(0) | | Authors: | Yu, X, Han, S. | | Deposit date: | 2018-10-05 | | Release date: | 2019-11-06 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Cryo-EM structures of the human glutamine transporter SLC1A5 (ASCT2) in the outward-facing conformation.
Elife, 8, 2019
|
|
