7C1S
 
 | | Crystal structure of the starter condensation domain of rhizomide synthetase RzmA mutant H140A/R148A in complex with C8-CoA and Leu-SNAC | | Descriptor: | Non-ribosomal peptide synthetase modules, OCTANOYL-COENZYME A, S-(2-acetamidoethyl) (2S)-2-azanyl-4-methyl-pentanethioate | | Authors: | Zhong, L, Diao, X, Zhang, N, Li, F.W, Zhou, H.B, Chen, H.N, Ren, X, Zhang, Y, Wu, D, Bian, X. | | Deposit date: | 2020-05-05 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.586 Å) | | Cite: | Engineering and elucidation of the lipoinitiation process in nonribosomal peptide biosynthesis.
Nat Commun, 12, 2021
|
|
6NBF
 
 | | Cryo-EM structure of parathyroid hormone receptor type 1 in complex with a long-acting parathyroid hormone analog and G protein | | Descriptor: | CHOLESTEROL, Gs protein alpha subunit, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhao, L.-H, Ma, S, Sutkeviciute, I, Shen, D.-D, Zhou, X.E, de Waal, P.P, Li, C.-Y, Kang, Y, Clark, L.J, Jean-Alphonse, F.G, White, A.D, Xiao, K, Yang, D, Jiang, Y, Watanabe, T, Gardella, T.J, Melcher, K, Wang, M.-W, Vilardaga, J.-P, Xu, H.E, Zhang, Y. | | Deposit date: | 2018-12-07 | | Release date: | 2019-04-17 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure and dynamics of the active human parathyroid hormone receptor-1.
Science, 364, 2019
|
|
5KPS
 
 | | Structure of RelA bound to ribosome in absence of A/R tRNA (Structure I) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Bah, E, Madireddy, R, Zhang, Y, Brilot, A.F, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2016-07-05 | | Release date: | 2016-09-28 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Ribosome•RelA structures reveal the mechanism of stringent response activation.
Elife, 5, 2016
|
|
7C1K
 
 | | Crystal structure of the starter condensation domain of rhizomide synthetase RzmA mutant R148A | | Descriptor: | Non-ribosomal peptide synthetase modules | | Authors: | Zhong, L, Diao, X, Zhang, N, Li, F.W, Zhou, H.B, Chen, H.N, Ren, X, Zhang, Y, Wu, D, Bian, X. | | Deposit date: | 2020-05-04 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.755 Å) | | Cite: | Engineering and elucidation of the lipoinitiation process in nonribosomal peptide biosynthesis.
Nat Commun, 12, 2021
|
|
7O6E
 
 | | 2.12 A cryo-EM structure of Mycobacterium tuberculosis Ferritin | | Descriptor: | Ferritin BfrB | | Authors: | Gijsbers, A, Zhang, Y, Gao, Y, Peters, P.J, Ravelli, R.B.G. | | Deposit date: | 2021-04-10 | | Release date: | 2021-05-19 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Mycobacterium tuberculosis ferritin: a suitable workhorse protein for cryo-EM development.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
1XHL
 
 | | Crystal Structure of putative Tropinone Reductase-II from Caenorhabditis Elegans with Cofactor and Substrate | | Descriptor: | 8-METHYL-8-AZABICYCLO[3,2,1]OCTAN-3-ONE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short-chain dehydrogenase/reductase family member (5L265), ... | | Authors: | Schormann, N, Karpova, E, Zhou, J, Zhang, Y, Symersky, J, Bunzel, R, Huang, W.-Y, Arabshahi, A, Qiu, S, Luan, C.-H, Gray, R, Carson, M, Tsao, J, Luo, M, Johnson, D, Lu, S, Lin, G, Luo, D, Cao, Z, Li, S, McKInstry, A, Shang, Q, Chen, Y.-J, Bray, T, Nagy, L, DeLucas, L, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-09-20 | | Release date: | 2004-09-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of putative Tropinone Reductase-II from Caenorhabditis Elegans with Cofactor and Substrate
To be Published
|
|
6O6M
 
 | | The Structure of EgtB (Cabther) | | Descriptor: | EgtB (Cabther), FE (III) ION, GLYCEROL | | Authors: | Irani, S, Zhang, Y. | | Deposit date: | 2019-03-07 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.506 Å) | | Cite: | Crystal Structure of the Ergothioneine Sulfoxide Synthase fromCandidatus Chloracidobacterium thermophilumand Structure-Guided Engineering To Modulate Its Substrate Selectivity.
Acs Catalysis, 9, 2019
|
|
4U7D
 
 | | Structure of human RECQ-like helicase in complex with an oligonucleotide | | Descriptor: | ATP-dependent DNA helicase Q1, DNA oligonucleotide, ZINC ION | | Authors: | Pike, A.C.W, Zhang, Y, Schnecke, C, Cooper, C.D.O, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-07-30 | | Release date: | 2015-01-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Human RECQ1 helicase-driven DNA unwinding, annealing, and branch migration: Insights from DNA complex structures.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6NBI
 
 | | Cryo-EM structure of parathyroid hormone receptor type 1 in complex with a long-acting parathyroid hormone analog and G protein | | Descriptor: | CHOLESTEROL, Gs protein alpha subunit, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhao, L.-H, Ma, S, Sutkeviciute, I, Shen, D.-D, Zhou, X.E, de Waal, P.P, Li, C.-Y, Kang, Y, Clark, L.J, Jean-Alphonse, F.G, White, A.D, Xiao, K, Yang, D, Jiang, Y, Watanabe, T, Gardella, T.J, Melcher, K, Wang, M.-W, Vilardaga, J.-P, Xu, H.E, Zhang, Y. | | Deposit date: | 2018-12-07 | | Release date: | 2019-04-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure and dynamics of the active human parathyroid hormone receptor-1.
Science, 364, 2019
|
|
5B7J
 
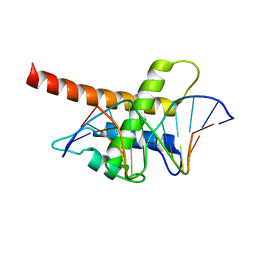 | | Structure model of Sap1-DNA complex | | Descriptor: | DNA (5'-D(*AP*AP*TP*AP*TP*TP*GP*TP*TP*TP*TP*G)-3'), DNA (5'-D(*CP*AP*AP*AP*AP*CP*AP*AP*TP*AP*TP*T)-3'), Switch-activating protein 1 | | Authors: | Jin, C, Hu, Y, Ding, J, Zhang, Y. | | Deposit date: | 2016-06-07 | | Release date: | 2017-02-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Sap1 is a replication-initiation factor essential for the assembly of pre-replicative complex in the fission yeast Schizosaccharomyces pombe
J. Biol. Chem., 292, 2017
|
|
3JCS
 
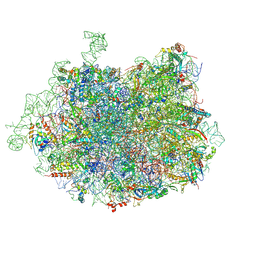 | | 2.8 Angstrom cryo-EM structure of the large ribosomal subunit from the eukaryotic parasite Leishmania | | Descriptor: | 26S alpha ribosomal RNA, 26S delta ribosomal RNA, 26S epsilon ribosomal RNA, ... | | Authors: | Shalev-Benami, M, Zhang, Y, Matzov, D, Halfon, Y, Zackay, A, Rozenberg, H, Zimmerman, E, Bashan, A, Jaffe, C.L, Yonath, A, Skiniotis, G. | | Deposit date: | 2016-01-21 | | Release date: | 2016-07-20 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | 2.8- angstrom Cryo-EM Structure of the Large Ribosomal Subunit from the Eukaryotic Parasite Leishmania.
Cell Rep, 16, 2016
|
|
1XKQ
 
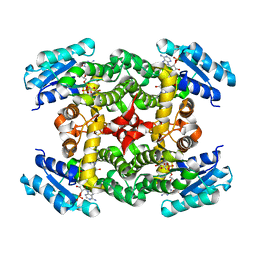 | | Crystal Structure of Short-Chain Dehydrogenase/Reductase of unknown Function from Caenorhabditis Elegans with Cofactor | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, short-chain reductase family member (5D234) | | Authors: | Schormann, N, Zhou, J, Karpova, E, Zhang, Y, Symersky, J, Bunzel, B, Huang, W.-Y, Arabshahi, A, Qiu, S, Luan, C.-H, Gray, R, Carson, M, Tsao, J, Luo, M, Johnson, D, Lu, S, Lin, G, Luo, D, Cao, Z, Li, S, McKinstry, A, Shang, Q, Chen, Y.-J, Bray, T, Nagy, L, DeLucas, L, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-09-29 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Short-Chain Dehydrogenase/Reductase of unknown Function from Caenorhabditis Elegans with Cofactor
To be Published
|
|
5WPW
 
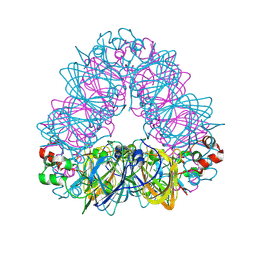 | | Crystal structure of coconut allergen cocosin | | Descriptor: | 11S globulin isoform 1 | | Authors: | Jin, T, Zhang, Y. | | Deposit date: | 2016-11-21 | | Release date: | 2017-10-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.847 Å) | | Cite: | Crystal Structure of Cocosin, A Potential Food Allergen from Coconut (Cocos nucifera)
J. Agric. Food Chem., 65, 2017
|
|
1YIS
 
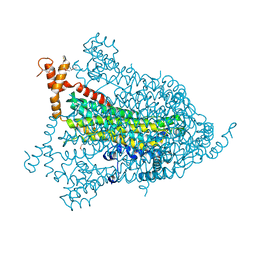 | | Structural genomics of Caenorhabditis elegans: adenylosuccinate lyase | | Descriptor: | SULFATE ION, adenylosuccinate lyase | | Authors: | Symersky, J, Schormann, N, Lu, S, Zhang, Y, Karpova, E, Qiu, S, Huang, W, Cao, Z, Zhou, J, Luo, M, Arabshahi, A, McKinstry, A, Luan, C.-H, Luo, D, Johnson, D, An, J, Tsao, J, Delucas, L, Shang, Q, Gray, R, Li, S, Bray, T, Chen, Y.-J, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2005-01-12 | | Release date: | 2005-01-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural genomics of Caenorhabditis elegans: adenylosuccinate lyase
To be Published
|
|
3L0B
 
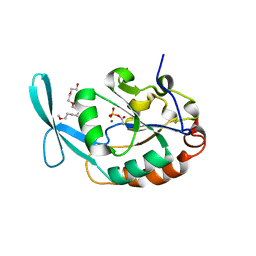 | | Crystal structure of SCP1 phosphatase D206A mutant phosphoryl-intermediate | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, Carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 1, MAGNESIUM ION | | Authors: | Zhang, M, Zhang, Y. | | Deposit date: | 2009-12-09 | | Release date: | 2010-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and functional analysis of the phosphoryl transfer reaction mediated by the human small C-terminal domain phosphatase, Scp1.
Protein Sci., 19, 2010
|
|
3L0Y
 
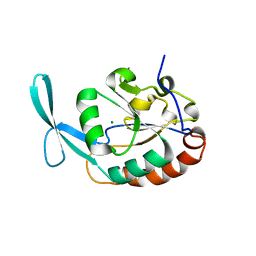 | | Crystal structure OF SCP1 phosphatase D98A mutant | | Descriptor: | Carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 1, MAGNESIUM ION | | Authors: | Zhang, M, Zhang, Y. | | Deposit date: | 2009-12-10 | | Release date: | 2010-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and functional analysis of the phosphoryl transfer reaction mediated by the human small C-terminal domain phosphatase, Scp1.
Protein Sci., 19, 2010
|
|
3L0C
 
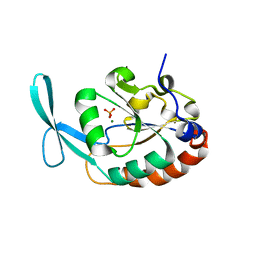 | |
2RFS
 
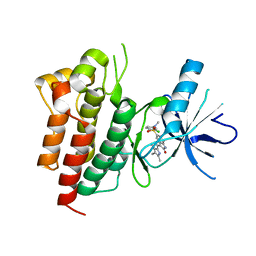 | | X-ray structure of SU11274 bound to c-Met | | Descriptor: | Hepatocyte growth factor receptor, N-(3-chlorophenyl)-N-methyl-2-oxo-3-[(3,4,5-trimethyl-1H-pyrrol-2-yl)methyl]-2H-indole-5-sulfonamide | | Authors: | Bellon, S.F, Kaplan-Lefko, P, Yang, Y, Zhang, Y, Moriguchi, J, Dussault, I. | | Deposit date: | 2007-10-01 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | c-Met inhibitors with novel binding mode show activity against several hereditary papillary renal cell carcinoma-related mutations.
J.Biol.Chem., 283, 2008
|
|
6V4X
 
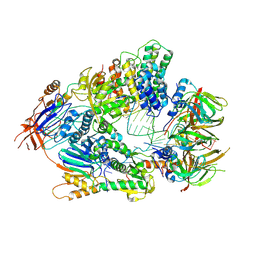 | | Cryo-EM structure of an active human histone pre-mRNA 3'-end processing machinery at 3.2 Angstrom resolution | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 2, Cleavage and polyadenylation specificity factor subunit 3, Small nuclear ribonucleoprotein E, ... | | Authors: | Sun, Y, Zhang, Y, Walz, T, Tong, L. | | Deposit date: | 2019-12-02 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of an active human histone pre-mRNA 3'-end processing machinery.
Science, 367, 2020
|
|
7SH6
 
 | |
4CKH
 
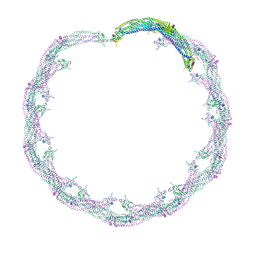 | | Helical reconstruction of ACAP1(BAR-PH domain) decorated membrane tubules by cryo-electron microscopy | | Descriptor: | ARF-GAP WITH COILED-COIL, ANK REPEAT AND PH DOMAIN-CONTAINING PROTEIN 1 | | Authors: | Pang, X.Y, Fan, J, Zhang, Y, Zhang, K, Gao, B.Q, Ma, J, Li, J, Deng, Y.C, Zhou, Q.J, Hsu, V, Sun, F. | | Deposit date: | 2014-01-06 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | A Ph Domain in Acap1 Possesses Key Features of the Bar Domain in Promoting Membrane Curvature.
Dev.Cell, 31, 2014
|
|
2RFN
 
 | | x-ray structure of c-Met with inhibitor. | | Descriptor: | 2-benzyl-5-(3-fluoro-4-{[6-methoxy-7-(3-morpholin-4-ylpropoxy)quinolin-4-yl]oxy}phenyl)-3-methylpyrimidin-4(3H)-one, Hepatocyte growth factor receptor | | Authors: | Bellon, S.F, Kaplan-Lefko, P, Yang, Y, Zhang, Y, Moriguchi, J, Dussault, I. | | Deposit date: | 2007-10-01 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | c-Met inhibitors with novel binding mode show activity against several hereditary papillary renal cell carcinoma-related mutations.
J.Biol.Chem., 283, 2008
|
|
6VX7
 
 | | bestrophin-2 Ca2+-bound state (5 mM Ca2+) | | Descriptor: | Bestrophin, CALCIUM ION, CHLORIDE ION | | Authors: | Owji, A.P, Zhao, Q, Ji, C, Kittredge, A, Hopiavuori, A, Fu, Z, Ward, N, Clarke, O, Shen, Y, Zhang, Y, Hendrickson, W.A, Yang, T. | | Deposit date: | 2020-02-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.36 Å) | | Cite: | Structural and functional characterization of the bestrophin-2 anion channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
5VNU
 
 | | Nonheme Iron Replacement in a Biosynthetic Nitric Oxide Reductase Model Performing O2 Reduction to Water: Mn-bound FeBMb | | Descriptor: | MANGANESE (II) ION, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Reed, J, Shi, Y, Zhu, Q, Chakraborty, S, Mirs, E.N, Petrik, I.D, Bhagi-Damodaran, A, Ross, M, Moenne-Loccoz, P, Zhang, Y, Lu, Y. | | Deposit date: | 2017-05-01 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.584 Å) | | Cite: | Manganese and Cobalt in the Nonheme-Metal-Binding Site of a Biosynthetic Model of Heme-Copper Oxidase Superfamily Confer Oxidase Activity through Redox-Inactive Mechanism.
J. Am. Chem. Soc., 139, 2017
|
|
7N4W
 
 | | Complex structure of ROTU4 with rotundine | | Descriptor: | GLYCEROL, ROTU4, SULFATE ION, ... | | Authors: | Kim, W, Zhang, Y. | | Deposit date: | 2021-06-04 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Using fungible biosensors to evolve improved alkaloid biosyntheses.
Nat.Chem.Biol., 18, 2022
|
|
