6LZE
 
 | | The crystal structure of COVID-19 main protease in complex with an inhibitor 11a | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, ~{N}-[(2~{S})-3-cyclohexyl-1-oxidanylidene-1-[[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]propan-2-yl]-1~{H}-indole-2-carboxamide | | 著者 | Zhang, B, Zhang, Y, Jing, Z, Liu, X, Yang, H, Liu, H, Rao, Z, Jiang, H. | | 登録日 | 2020-02-19 | | 公開日 | 2020-04-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.505 Å) | | 主引用文献 | Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease.
Science, 368, 2020
|
|
7AW0
 
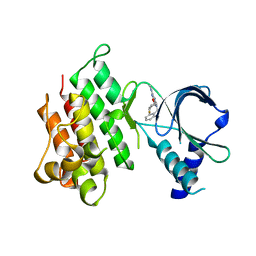 | | MerTK kinase domain in complex with purine inhibitor | | 分子名称: | 2-(cyclopentyloxy)-9-(2,6-difluorobenzyl)-N-methyl-9H-purin-6-amine, Tyrosine-protein kinase Mer | | 著者 | Schimpl, M, Nissink, J.W.M, Blackett, C, Clarke, M, Disch, J, Goldberg, K, Guilinger, J, Hennessy, E.J, Jetson, R, Ginkunja, D, Hardaker, E, Keefe, A, McCoull, W, McMurray, L, Collingwood, O, Overman, R, Pflug, A, Preston, M, Rawlins, P, Rivers, E, Smith, P, Underwood, E, Truman, C, Warwicker, J, Winter, J, Woodcock, S, Zhang, Y. | | 登録日 | 2020-11-06 | | 公開日 | 2021-03-03 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.893 Å) | | 主引用文献 | Generating Selective Leads for Mer Kinase Inhibitors-Example of a Comprehensive Lead-Generation Strategy.
J.Med.Chem., 64, 2021
|
|
5KPS
 
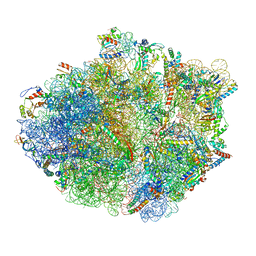 | | Structure of RelA bound to ribosome in absence of A/R tRNA (Structure I) | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Loveland, A.B, Bah, E, Madireddy, R, Zhang, Y, Brilot, A.F, Grigorieff, N, Korostelev, A.A. | | 登録日 | 2016-07-05 | | 公開日 | 2016-09-28 | | 最終更新日 | 2019-11-20 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Ribosome•RelA structures reveal the mechanism of stringent response activation.
Elife, 5, 2016
|
|
6V4P
 
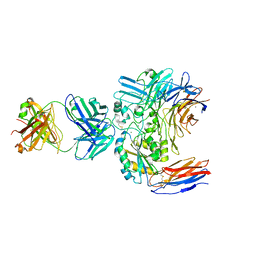 | | Structure of the integrin AlphaIIbBeta3-Abciximab complex | | 分子名称: | Abciximab, heavy chain, light chain, ... | | 著者 | Nesic, D, Zhang, Y, Spasic, A, Li, J, Provasi, D, Filizola, M, Walz, T, Coller, B.S. | | 登録日 | 2019-11-28 | | 公開日 | 2020-02-05 | | 最終更新日 | 2020-03-11 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Cryo-Electron Microscopy Structure of the alpha IIb beta 3-Abciximab Complex.
Arterioscler Thromb Vasc Biol., 40, 2020
|
|
5IMQ
 
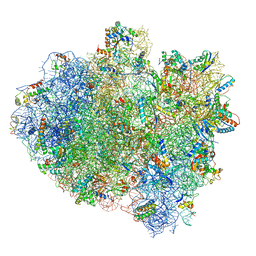 | | Structure of ribosome bound to cofactor at 3.8 angstrom resolution | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Kumar, V, Ero, R, Jian, G.K, Ahmed, T, Zhan, Y, Bhushan, S, Gao, Y.G. | | 登録日 | 2016-03-06 | | 公開日 | 2016-05-18 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structure of the GTP Form of Elongation Factor 4 (EF4) Bound to the Ribosome
J.Biol.Chem., 291, 2016
|
|
5EZ6
 
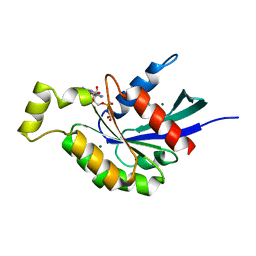 | | Crystallization and preliminary X-ray crystallographic analysis of a small GTPase RhoA | | 分子名称: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Transforming protein RhoA | | 著者 | Yan, Z, Ma, S, Zhang, Y, Ma, L, Wang, F, Li, J, Miao, L. | | 登録日 | 2015-11-26 | | 公開日 | 2016-12-07 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystallization and preliminary X-ray crystallographic analysis of a small GTPase RhoA
To Be Published
|
|
6VG7
 
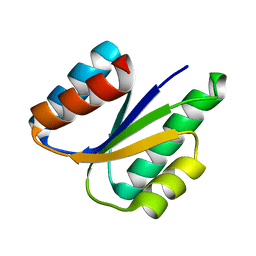 | |
4EAY
 
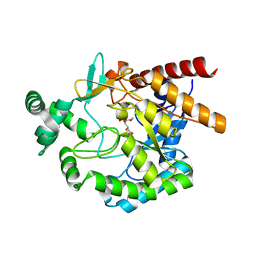 | | Crystal structures of mannonate dehydratase from Escherichia coli strain K12 complexed with D-mannonate | | 分子名称: | CHLORIDE ION, D-MANNONIC ACID, MANGANESE (II) ION, ... | | 著者 | Qiu, X, Zhu, Y, Yuan, Y, Zhang, Y, Liu, H, Gao, Y, Teng, M, Niu, L. | | 登録日 | 2012-03-23 | | 公開日 | 2013-03-27 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structural insights into decreased enzymatic activity induced by an insert sequence in mannonate dehydratase from Gram negative bacterium.
J.Struct.Biol., 180, 2012
|
|
7W41
 
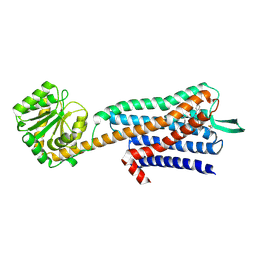 | |
1P58
 
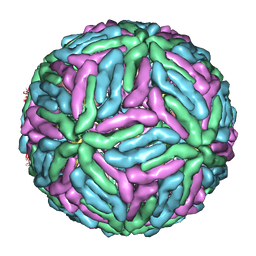 | | Complex Organization of Dengue Virus Membrane Proteins as Revealed by 9.5 Angstrom Cryo-EM reconstruction | | 分子名称: | Envelope protein M, Major envelope protein E | | 著者 | Zhang, W, Chipman, P.R, Corver, J, Johnson, P.R, Zhang, Y, Mukhopadhyay, S, Baker, T.S, Strauss, J.H, Rossmann, M.G, Kuhn, R.J. | | 登録日 | 2003-04-25 | | 公開日 | 2003-11-04 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (9.5 Å) | | 主引用文献 | Visualization of membrane protein domains by cryo-electron microscopy of dengue virus
Nat.Struct.Biol., 10, 2003
|
|
6VGA
 
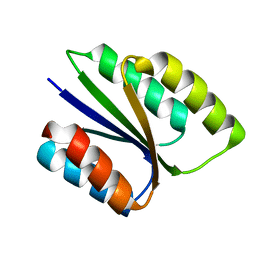 | |
6VGB
 
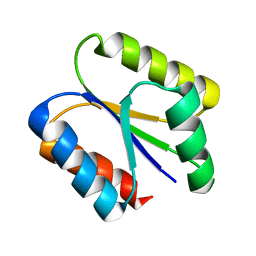 | |
3J8G
 
 | | Electron cryo-microscopy structure of EngA bound with the 50S ribosomal subunit | | 分子名称: | 23S rRNA, 50S ribosomal protein L1, 50S ribosomal protein L11, ... | | 著者 | Zhang, X, Yan, K, Zhang, Y, Li, N, Ma, C, Li, Z, Zhang, Y, Feng, B, Liu, J, Sun, Y, Xu, Y, Lei, J, Gao, N. | | 登録日 | 2014-10-24 | | 公開日 | 2014-11-26 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (5 Å) | | 主引用文献 | Structural insights into the function of a unique tandem GTPase EngA in bacterial ribosome assembly
Nucleic Acids Res., 2014
|
|
5U0P
 
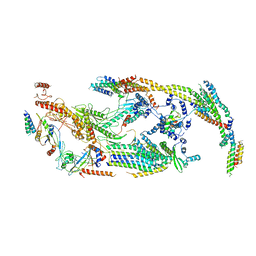 | | Cryo-EM structure of the transcriptional Mediator | | 分子名称: | Mediator complex subunit 10, Mediator complex subunit 11, Mediator complex subunit 14, ... | | 著者 | Tsai, K.-L, Yu, X, Gopalan, S, Chao, T.-C, Zhang, Y, Florens, L, Washburn, M.P, Murakami, K, Conaway, R.C, Conaway, J.W, Asturias, F. | | 登録日 | 2016-11-26 | | 公開日 | 2017-03-08 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Mediator structure and rearrangements required for holoenzyme formation.
Nature, 544, 2017
|
|
5U9D
 
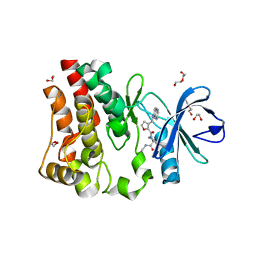 | | Discovery of a potent BTK inhibitor with a novel binding mode using parallel selections with a DNA-encoded chemical library | | 分子名称: | (R)-N-methyl-2-(3-((quinoxalin-6-ylamino)methyl)furan-2-carbonyl)-2,3,4,9-tetrahydro-1H-pyrido[3,4-b]indole-3-carboxamide, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Cuozzo, J.W, Centrella, P.A, Gikunju, D, Habeshian, S, Hupp, C.D, Keefe, A.D, Sigel, E, Soutter, H.H, Thomson, H.A, Zhang, Y, Clark, M.A. | | 登録日 | 2016-12-16 | | 公開日 | 2017-01-18 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.33 Å) | | 主引用文献 | Discovery of a Potent BTK Inhibitor with a Novel Binding Mode by Using Parallel Selections with a DNA-Encoded Chemical Library.
Chembiochem, 18, 2017
|
|
3CIV
 
 | |
4EAC
 
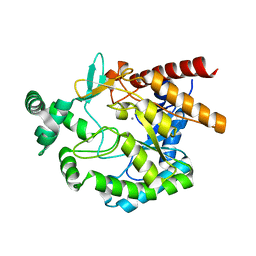 | | Crystal structure of mannonate dehydratase from Escherichia coli strain K12 | | 分子名称: | CHLORIDE ION, MANGANESE (II) ION, Mannonate dehydratase | | 著者 | Qiu, X, Zhu, Y, Yuan, Y, Zhang, Y, Liu, H, Gao, Y, Teng, M, Niu, L. | | 登録日 | 2012-03-22 | | 公開日 | 2013-03-27 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural insights into decreased enzymatic activity induced by an insert sequence in mannonate dehydratase from Gram negative bacterium.
J.Struct.Biol., 180, 2012
|
|
5U0S
 
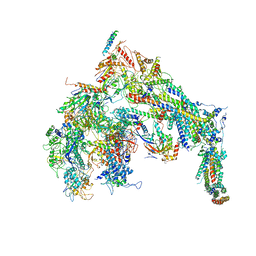 | | Cryo-EM structure of the Mediator-RNAPII complex | | 分子名称: | Mediator complex subunit 10, Mediator complex subunit 11, Mediator complex subunit 14, ... | | 著者 | Tsai, K.-L, Yu, X, Gopalan, S, Chao, T.-C, Zhang, Y, Florens, L, Washburn, M.P, Murakami, K, Conaway, R.C, Conaway, J.W, Asturias, F. | | 登録日 | 2016-11-26 | | 公開日 | 2017-03-08 | | 最終更新日 | 2020-01-01 | | 実験手法 | ELECTRON MICROSCOPY (7.8 Å) | | 主引用文献 | Mediator structure and rearrangements required for holoenzyme formation.
Nature, 544, 2017
|
|
1K4V
 
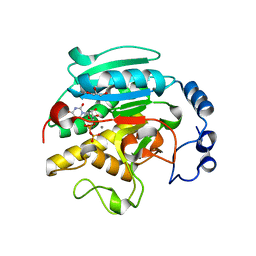 | | 1.53 A Crystal Structure of the Beta-Galactoside-alpha-1,3-galactosyltransferase in Complex with UDP | | 分子名称: | GLYCEROL, MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, ... | | 著者 | Boix, E, Swaminathan, G.J, Zhang, Y, Natesh, R, Brew, K, Acharya, K.R. | | 登録日 | 2001-10-09 | | 公開日 | 2002-04-10 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.53 Å) | | 主引用文献 | Structure of UDP complex of UDP-galactose:beta-galactoside-alpha -1,3-galactosyltransferase at 1.53-A resolution reveals a conformational change in the catalytically important C terminus.
J.Biol.Chem., 276, 2001
|
|
5IMR
 
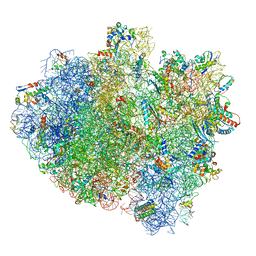 | | Structure of ribosome bound to cofactor at 5.7 angstrom resolution | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Kumar, V, Ero, R, Jian, G.K, Ahmed, T, Zhan, Y, Bhushan, S, Gao, Y.G. | | 登録日 | 2016-03-06 | | 公開日 | 2016-05-18 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (5.7 Å) | | 主引用文献 | Structure of the GTP Form of Elongation Factor 4 (EF4) Bound to the Ribosome
J.Biol.Chem., 291, 2016
|
|
6VLS
 
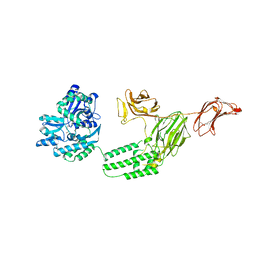 | | Structure of C-terminal fragment of Vip3A toxin | | 分子名称: | DI(HYDROXYETHYL)ETHER, Maltose/maltodextrin-binding periplasmic protein,Vip3Aa | | 著者 | Jiang, K, Zhang, Y, Chen, Z, Gao, X. | | 登録日 | 2020-01-25 | | 公開日 | 2020-07-22 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structural and Functional Insights into the C-terminal Fragment of Insecticidal Vip3A Toxin ofBacillus thuringiensis.
Toxins, 12, 2020
|
|
5UZ7
 
 | | Volta phase plate cryo-electron microscopy structure of a calcitonin receptor-heterotrimeric Gs protein complex | | 分子名称: | Calcitonin receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Liang, Y.L, Khoshouei, M, Radjainia, M, Zhang, Y, Glukhova, A, Tarrasch, J, Thal, D.M, Furness, S.G.B, Christopoulos, G, Coudrat, T, Danev, R, Baumeister, W, Miller, L.J, Christopoulos, A, Kobilka, B.K, Wootten, D, Skiniotis, G, Sexton, P.M. | | 登録日 | 2017-02-24 | | 公開日 | 2017-05-03 | | 最終更新日 | 2018-07-18 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Phase-plate cryo-EM structure of a class B GPCR-G-protein complex.
Nature, 546, 2017
|
|
4TKQ
 
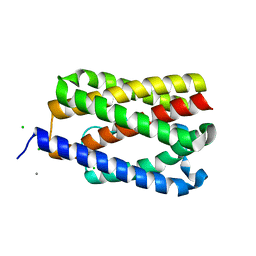 | | Native-SAD phasing for YetJ from Bacillus Subtilis | | 分子名称: | CALCIUM ION, CHLORIDE ION, Uncharacterized protein YetJ | | 著者 | Liu, Q, Chang, Y, Hendrickson, W.A, New York Consortium on Membrane Protein Structure (NYCOMPS) | | 登録日 | 2014-05-27 | | 公開日 | 2014-06-18 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.8025 Å) | | 主引用文献 | Multi-crystal native SAD analysis at 6 keV.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3ORW
 
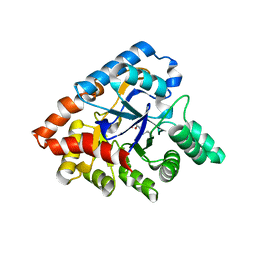 | | Crystal structure of thermophilic phosphotriesterase from Geobacillus kaustophilus HTA426 | | 分子名称: | COBALT (II) ION, Phosphotriesterase | | 著者 | Zheng, B.S, Yu, S.S, Zhang, Y, Lou, Z.Y, Feng, Y. | | 登録日 | 2010-09-07 | | 公開日 | 2011-09-21 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of thermophilic phosphotriesterase from Geobacillus kaustophilus HTA426
To be Published
|
|
4CSU
 
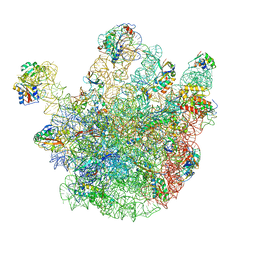 | | Cryo-EM structures of the 50S ribosome subunit bound with ObgE | | 分子名称: | 23S RRNA, 50S RIBOSOMAL PROTEIN L1, 50S RIBOSOMAL PROTEIN L11, ... | | 著者 | Feng, B, Mandava, C.S, Guo, Q, Wang, J, Cao, W, Li, N, Zhang, Y, Zhang, Y, Wang, Z, Wu, J, Sanyal, S, Lei, J, Gao, N. | | 登録日 | 2014-03-10 | | 公開日 | 2014-06-04 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (5.5 Å) | | 主引用文献 | Structural and Functional Insights Into the Mode of Action of a Universally Conserved Obg Gtpase.
Plos Biol., 12, 2014
|
|
