3RC0
 
 | | Human SETD6 in complex with RelA Lys310 peptide | | 分子名称: | 1,2-ETHANEDIOL, N-lysine methyltransferase SETD6, S-ADENOSYLMETHIONINE, ... | | 著者 | Chang, Y, Levy, D, Horton, J.R, Peng, J, Zhang, X, Gozani, O, Cheng, X. | | 登録日 | 2011-03-30 | | 公開日 | 2011-05-25 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | Structural basis of SETD6-mediated regulation of the NF-kB network via methyl-lysine signaling.
Nucleic Acids Res., 39, 2011
|
|
6UTU
 
 | | Crystal structure of minor pseudopilin ternary complex of XcpVWX from the Type 2 secretion system of Pseudomonas aeruginosa in the P3 space group | | 分子名称: | CALCIUM ION, Type II secretion system protein I, Type II secretion system protein J, ... | | 著者 | Zhang, Y, Wang, S, Jia, Z. | | 登録日 | 2019-10-30 | | 公開日 | 2020-09-02 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | In Situ Proteolysis Condition-Induced Crystallization of the XcpVWX Complex in Different Lattices.
Int J Mol Sci, 21, 2020
|
|
3QXY
 
 | | Human SETD6 in complex with RelA Lys310 | | 分子名称: | 1,2-ETHANEDIOL, N-lysine methyltransferase SETD6, S-ADENOSYLMETHIONINE, ... | | 著者 | Chang, Y, Levy, D, Horton, J.R, Peng, J, Zhang, X, Gozani, O, Cheng, X. | | 登録日 | 2011-03-02 | | 公開日 | 2011-05-25 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Structural basis of SETD6-mediated regulation of the NF-kB network via methyl-lysine signaling.
Nucleic Acids Res., 39, 2011
|
|
8CZJ
 
 | | A bacteria Zrt/Irt-like protein in the apo state | | 分子名称: | Putative membrane protein, SULFATE ION, [(Z)-octadec-9-enyl] (2R)-2,3-bis(oxidanyl)propanoate | | 著者 | Zhang, Y, Hu, J. | | 登録日 | 2022-05-24 | | 公開日 | 2023-02-22 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.751 Å) | | 主引用文献 | Structural insights into the elevator-type transport mechanism of a bacterial ZIP metal transporter.
Nat Commun, 14, 2023
|
|
3OMW
 
 | | Crystal structure of Ssu72, an essential eukaryotic phosphatase specific for the C-terminal domain of RNA polymerase II | | 分子名称: | CG14216 | | 著者 | Zhang, Y, Zhang, M, Zhang, Y. | | 登録日 | 2010-08-27 | | 公開日 | 2011-01-19 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.8701 Å) | | 主引用文献 | Crystal structure of Ssu72, an essential eukaryotic phosphatase specific for the C-terminal domain of RNA polymerase II, in complex with a transition state analogue.
Biochem.J., 434, 2011
|
|
8E4V
 
 | |
7BZ2
 
 | | Cryo-EM structure of the formoterol-bound beta2 adrenergic receptor-Gs protein complex. | | 分子名称: | Beta2 adrenergic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Zhang, Y.N, Yang, F, Ling, S.L, Lv, P, Zhou, Y.X, Fang, W, Sun, W, Shi, P, Tian, C.L. | | 登録日 | 2020-04-26 | | 公開日 | 2020-08-05 | | 実験手法 | ELECTRON MICROSCOPY (3.82 Å) | | 主引用文献 | Single-particle cryo-EM structural studies of the beta2AR-Gs complex bound with a full agonist formoterol.
Cell Discov, 6, 2020
|
|
3QO2
 
 | | Structural insights for MPP8 chromodomain interaction with histone H3 lysine 9 | | 分子名称: | 1,2-ETHANEDIOL, Histone H3 peptide, M-phase phosphoprotein 8 | | 著者 | Chang, Y, Horton, J.R, Bedford, M.T, Zhang, X, Cheng, X. | | 登録日 | 2011-02-09 | | 公開日 | 2011-04-06 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.49 Å) | | 主引用文献 | Structural insights for MPP8 chromodomain interaction with histone H3 lysine 9: potential effect of phosphorylation on methyl-lysine binding.
J.Mol.Biol., 408, 2011
|
|
3OG7
 
 | | B-Raf Kinase V600E oncogenic mutant in complex with PLX4032 | | 分子名称: | AKAP9-BRAF fusion protein, N-(3-{[5-(4-chlorophenyl)-1H-pyrrolo[2,3-b]pyridin-3-yl]carbonyl}-2,4-difluorophenyl)propane-1-sulfonamide | | 著者 | Zhang, Y, Zhang, K.Y, Zhang, C. | | 登録日 | 2010-08-16 | | 公開日 | 2010-09-22 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma.
Nature, 467, 2010
|
|
3OMX
 
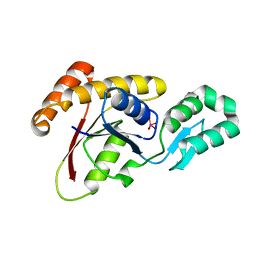 | | Crystal structure of Ssu72 with vanadate complex | | 分子名称: | CG14216, VANADATE ION | | 著者 | Zhang, Y, Zhang, M, Zhang, Y. | | 登録日 | 2010-08-27 | | 公開日 | 2011-01-19 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.3366 Å) | | 主引用文献 | Crystal structure of Ssu72, an essential eukaryotic phosphatase specific for the C-terminal domain of RNA polymerase II, in complex with a transition state analogue.
Biochem.J., 434, 2011
|
|
5GSN
 
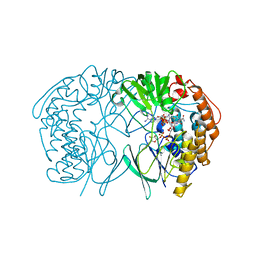 | | Tmm in complex with methimazole | | 分子名称: | 1-METHYL-1,3-DIHYDRO-2H-IMIDAZOLE-2-THIONE, FLAVIN-ADENINE DINUCLEOTIDE, Flavin-containing monooxygenase, ... | | 著者 | Zhang, Y.Z, Li, C.Y. | | 登録日 | 2016-08-16 | | 公開日 | 2017-01-18 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.203 Å) | | 主引用文献 | Structural mechanism for bacterial oxidation of oceanic trimethylamine into trimethylamine N-oxide
Mol. Microbiol., 103, 2017
|
|
1N6G
 
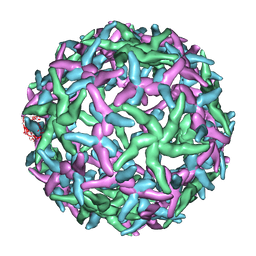 | | The structure of immature Dengue-2 prM particles | | 分子名称: | major envelope protein E | | 著者 | Zhang, Y, Corver, J, Chipman, P.R, Zhang, W, Pletnev, S.V, Sedlak, D, Baker, T.S, Strauss, J.H, Kuhn, R.J, Rossmann, M.G. | | 登録日 | 2002-11-10 | | 公開日 | 2003-06-03 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (16 Å) | | 主引用文献 | Structures of Immature flavivirus particles
EMBO J., 22, 2003
|
|
1NA4
 
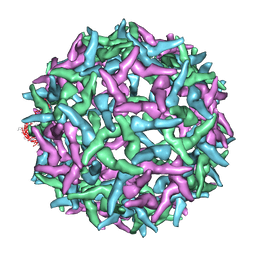 | | The structure of immature Yellow Fever virus particle | | 分子名称: | major envelope protein E | | 著者 | Zhang, Y, Corver, J, Chipman, P.R, Lenches, E, Zhang, W, Pletnev, S.V, Sedlak, D, Baker, T.S, Strauss, J.H, Kuhn, R.J, Rossmann, M.G. | | 登録日 | 2002-11-26 | | 公開日 | 2003-12-09 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY | | 主引用文献 | Structures of immature flavivirus particles
EMBO J., 22, 2003
|
|
7D0A
 
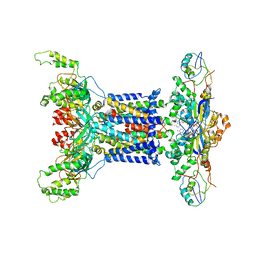 | | Acinetobacter MlaFEDB complex in ADP-vanadate trapped Vclose conformation | | 分子名称: | ABC transporter ATP-binding protein, ADENOSINE-5'-DIPHOSPHATE, Anti-sigma factor antagonist, ... | | 著者 | Zhang, Y.Y, Fan, Q.X, Chi, X.M, Zhou, Q, Li, Y.Y. | | 登録日 | 2020-09-09 | | 公開日 | 2020-12-16 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Cryo-EM structures of Acinetobacter baumannii glycerophospholipid transporter.
Cell Discov, 6, 2020
|
|
7D09
 
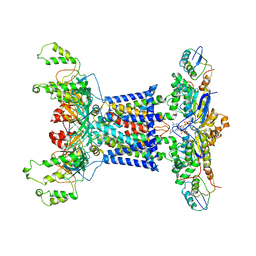 | | Acinetobacter MlaFEDB complex in ATP-bound Vtrans2 conformation | | 分子名称: | ABC transporter ATP-binding protein, ADENOSINE-5'-TRIPHOSPHATE, Anti-sigma factor antagonist, ... | | 著者 | Zhang, Y.Y, Fan, Q.X, Chi, X.M, Zhou, Q, Li, Y.Y. | | 登録日 | 2020-09-09 | | 公開日 | 2020-12-16 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Cryo-EM structures of Acinetobacter baumannii glycerophospholipid transporter.
Cell Discov, 6, 2020
|
|
1VZT
 
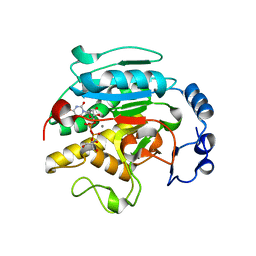 | | ROLES OF INDIVIDUAL RESIDUES OF ALPHA-1,3 GALACTOSYLTRANSFERASES IN SUBSTRATE BINDING AND CATALYSIS | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, ... | | 著者 | Zhang, Y, Swaminathan, G.J, Deshpande, A, Natesh, R, Xie, X, Acharya, K.R, Brew, K. | | 登録日 | 2004-05-26 | | 公開日 | 2005-05-26 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Roles of Individual Enzyme-Substrate Interactions by Alpha-1,3-Galactosyltransferase in Catalysis and Specificity.
Biochemistry, 42, 2003
|
|
5E1R
 
 | |
8JOO
 
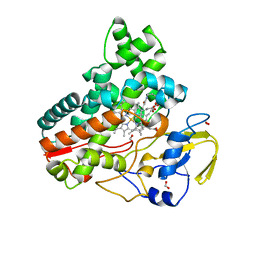 | | Crystal structure of cytochrome P450 IkaD from Streptomyces sp. ZJ306, in complex with the substrate ikarugamycin | | 分子名称: | (1Z,3E,5S,7R,8R,10R,11R,12S,15R,16S,18Z,25S)-11-ethyl-2-hydroxy-10-methyl-21,26-diazapentacyclo[23.2.1.05,16.07,15.08,12]octacosa-1(2),3,13,18-tetraene-20,27,28-trione, Cytochrome P450, FORMIC ACID, ... | | 著者 | Zhang, Y.L, Zhang, L.P, Zhang, C.S. | | 登録日 | 2023-06-08 | | 公開日 | 2023-11-15 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | A Mechanistic Understanding of the Distinct Regio- and Chemoselectivity of Multifunctional P450s by Structural Comparison of IkaD and CftA Complexed with Common Substrates.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
2ZUC
 
 | |
4DC2
 
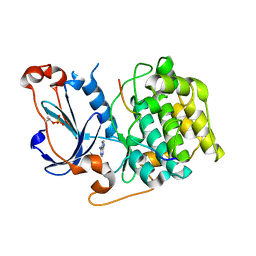 | | Structure of PKC in Complex with a Substrate Peptide from Par-3 | | 分子名称: | ADENINE, Partitioning defective 3 homolog, Protein kinase C iota type | | 著者 | Shang, Y, Wang, C, Yu, J, Zhang, M. | | 登録日 | 2012-01-17 | | 公開日 | 2012-07-11 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Substrate recognition mechanism of atypical protein kinase Cs revealed by the structure of PKC iota in complex with a substrate peptide from Par-3
Structure, 20, 2012
|
|
1PQN
 
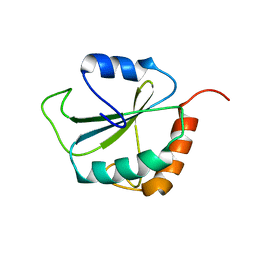 | | dominant negative human hDim1 (hDim1 1-128) | | 分子名称: | Spliceosomal U5 snRNP-specific 15 kDa protein | | 著者 | Zhang, Y.Z, Cheng, H, Gould, K.L, Golemis, E.A, Roder, H. | | 登録日 | 2003-06-18 | | 公開日 | 2003-08-26 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure, stability and function of hDim1 investigated by NMR, circular
dichroism and mutational analysis
Biochemistry, 42, 2003
|
|
7F7G
 
 | | a linear Peptide Inhibitors in complex with GK domain | | 分子名称: | DLG4 GK domain, UNK-ARG-ILE-ARG-ARG-ASP-GLU-TYR-LEU-LYS-ALA-ILE-GLN-UNK | | 著者 | Shang, Y, Huang, X, Li, X, Zhang, M. | | 登録日 | 2021-06-29 | | 公開日 | 2022-02-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.446 Å) | | 主引用文献 | Entropy of stapled peptide inhibitors in free state is the major contributor to the improvement of binding affinity with the GK domain.
Rsc Chem Biol, 2, 2021
|
|
3OPW
 
 | | Crystal Structure of the Rph1 catalytic core | | 分子名称: | DNA damage-responsive transcriptional repressor RPH1 | | 著者 | Chang, Y, Wu, J, Tong, X, Zhou, J, Ding, J. | | 登録日 | 2010-09-02 | | 公開日 | 2010-12-22 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of the catalytic core of Saccharomyces cerevesiae histone demethylase Rph1: insights into the substrate specificity and catalytic mechanism
Biochem.J., 433, 2011
|
|
7F7I
 
 | | Stapled Peptide Inhibitor in complex with PSD95 GK domain | | 分子名称: | ACE-ARG-ILE-ARG-ARG-ASP-GLU-TYR-LEU-LYZ-ALA-ILE-GLN-NH2, Disks large homolog 4 | | 著者 | Shang, Y, Huang, X, Li, X, Zhang, M. | | 登録日 | 2021-06-29 | | 公開日 | 2022-04-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.595 Å) | | 主引用文献 | Entropy of stapled peptide inhibitors in free state is the major contributor to the improvement of binding affinity with the GK domain.
Rsc Chem Biol, 2, 2021
|
|
3OPT
 
 | | Crystal structure of the Rph1 catalytic core with a-ketoglutarate | | 分子名称: | 2-OXOGLUTARIC ACID, DNA damage-responsive transcriptional repressor RPH1, NICKEL (II) ION | | 著者 | Chang, Y, Wu, J, Tong, X, Zhou, J, Ding, J. | | 登録日 | 2010-09-02 | | 公開日 | 2010-12-22 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of the catalytic core of Saccharomyces cerevesiae histone demethylase Rph1: insights into the substrate specificity and catalytic mechanism
Biochem.J., 433, 2011
|
|
