7W1L
 
 | | Crystal structure of carboxylesterase mutant from Thermobifida fusca with C8X | | Descriptor: | Carboxylesterase, bis(2-hydroxyethyl) benzene-1,4-dicarboxylate | | Authors: | Han, X, Gerlis, H, Li, Z, Gao, J, Wei, R, Liu, W. | | Deposit date: | 2021-11-19 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structural Insights into (Tere)phthalate-Ester Hydrolysis by a Carboxylesterase and Its Role in Promoting PET Depolymerization
Acs Catalysis, 12, 2022
|
|
7YKO
 
 | | Crystal structure of a novel alpha/beta hydrolase mutant from thermomonospora curvata in complex with pentane-1,5-diol | | Descriptor: | Triacylglycerol lipase, pentane-1,5-diol | | Authors: | Han, X, Jian, G, Bornscheuer, U.T, Wei, R, Liu, W. | | Deposit date: | 2022-07-23 | | Release date: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal structure of a novel alpha/beta hydrolase mutant from thermomonospora curvata in complex with pentane-1,5-diol
To Be Published
|
|
7YKP
 
 | | Crystal structure of a novel alpha/beta hydrolase from thermomonospora curvata with glycerol | | Descriptor: | GLYCEROL, Triacylglycerol lipase | | Authors: | Han, X, Jian, G, Bornscheuer, U.T, Wei, R, Liu, W. | | Deposit date: | 2022-07-23 | | Release date: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Crystal structure of a novel alpha/beta hydrolase from thermomonospora curvata with glycerol
To Be Published
|
|
7YKQ
 
 | |
8HK1
 
 | | The cryo-EM structure of human pre-17S U2 snRNP | | Descriptor: | ATP-dependent RNA helicase DDX42, HIV Tat-specific factor 1, PHD finger-like domain-containing protein 5A, ... | | Authors: | Zhang, X, Zhan, X, Shi, Y. | | Deposit date: | 2022-11-24 | | Release date: | 2023-03-08 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Mechanisms of the RNA helicases DDX42 and DDX46 in human U2 snRNP assembly.
Nat Commun, 14, 2023
|
|
8HMY
 
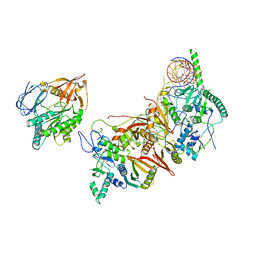 | | Cryo-EM structure of the human pre-catalytic TSEN/pre-tRNA complex | | Descriptor: | Chromosome 1 open reading frame 19, isoform CRA_a, MAGNESIUM ION, ... | | Authors: | Zhang, X, Yang, F, Zhan, X, Shi, Y. | | Deposit date: | 2022-12-06 | | Release date: | 2023-04-19 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Structural basis of pre-tRNA intron removal by human tRNA splicing endonuclease.
Mol.Cell, 83, 2023
|
|
8HMZ
 
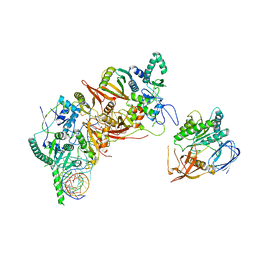 | | Cryo-EM structure of the human post-catalytic TSEN/pre-tRNA complex | | Descriptor: | Chromosome 1 open reading frame 19, isoform CRA_a, MAGNESIUM ION, ... | | Authors: | Zhang, X, Yang, F, Zhan, X, Shi, Y. | | Deposit date: | 2022-12-06 | | Release date: | 2023-04-19 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of pre-tRNA intron removal by human tRNA splicing endonuclease.
Mol.Cell, 83, 2023
|
|
1F3L
 
 | |
1PEG
 
 | | Structural basis for the product specificity of histone lysine methyltransferases | | Descriptor: | Histone H3, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION, ... | | Authors: | Zhang, X, Yang, Z, Khan, S.I, Horton, J.R, Tamaru, H, Selker, E.U, Cheng, X. | | Deposit date: | 2003-05-21 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural basis for the product specificity of histone lysine methyltransferases
Mol.Cell, 12, 2003
|
|
3JB7
 
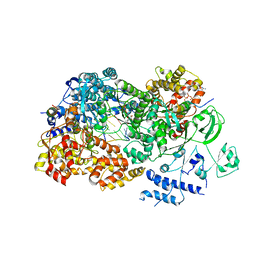 | | In situ structures of the segmented genome and RNA polymerase complex inside a dsRNA virus | | Descriptor: | CPV RNA-dependent RNA polymerase, CYTIDINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Zhang, X, Ding, K, Yu, X.K, Chang, W, Sun, J.C, Zhou, Z.H. | | Deposit date: | 2015-08-03 | | Release date: | 2015-10-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | In situ structures of the segmented genome and RNA polymerase complex inside a dsRNA virus.
Nature, 527, 2015
|
|
3JB6
 
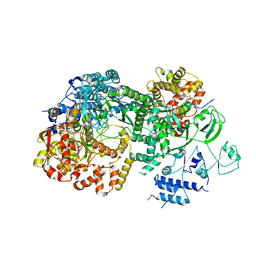 | | In situ structures of the segmented genome and RNA polymerase complex inside a dsRNA virus | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, RNA-dependent RNA polymerase, VP1 CSP, ... | | Authors: | Zhang, X, Ding, K, Yu, X.K, Chang, W, Sun, J.C, Zhou, Z.H. | | Deposit date: | 2015-08-02 | | Release date: | 2015-10-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | In situ structures of the segmented genome and RNA polymerase complex inside a dsRNA virus.
Nature, 527, 2015
|
|
1HQK
 
 | | CRYSTAL STRUCTURE ANALYSIS OF LUMAZINE SYNTHASE FROM AQUIFEX AEOLICUS | | Descriptor: | 6,7-DIMETHYL-8-RIBITYLLUMAZINE SYNTHASE | | Authors: | Zhang, X, Meining, W, Fischer, M, Bacher, A, Ladenstein, R. | | Deposit date: | 2000-12-18 | | Release date: | 2001-12-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray structure analysis and crystallographic refinement of lumazine synthase from the hyperthermophile Aquifex aeolicus at 1.6 A resolution: determinants of thermostability revealed from structural comparisons.
J.Mol.Biol., 306, 2001
|
|
1H0C
 
 | | The crystal structure of human alanine:glyoxylate aminotransferase | | Descriptor: | (AMINOOXY)ACETIC ACID, GLYCEROL, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Zhang, X, Danpure, C.J, Roe, S.M, Pearl, L.H. | | Deposit date: | 2002-06-17 | | Release date: | 2003-06-12 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Alanine:Glyoxylate Aminotransferase and the Relationship between Genotype and Enzymatic Phenotype in Primary Hyperoxaluria Type 1.
J.Mol.Biol., 331, 2003
|
|
6AHD
 
 | | The Cryo-EM Structure of Human Pre-catalytic Spliceosome (B complex) at 3.8 angstrom resolution | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, Brr2, U5 small nuclear ribonucleoprotein 200 kDa helicase, ... | | Authors: | Zhan, X, Yan, C, Zhang, X, Shi, Y. | | Deposit date: | 2018-08-17 | | Release date: | 2018-11-14 | | Last modified: | 2021-06-30 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of the human pre-catalytic spliceosome and its precursor spliceosome.
Cell Res., 28, 2018
|
|
8G05
 
 | | Cryo-EM structure of an orphan GPCR-Gi protein signaling complex | | Descriptor: | 6-(octylamino)pyrimidine-2,4(3H,5H)-dione, CHOLESTEROL, G-protein coupled receptor 84, ... | | Authors: | Zhang, X, Wang, Y.J, Li, X, Liu, G.B, Gong, W.M, Zhang, C. | | Deposit date: | 2023-01-31 | | Release date: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Pro-phagocytic function and structural basis of GPR84 signaling.
Nat Commun, 14, 2023
|
|
8H10
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Locked-2 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-10-19 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H14
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x3 Disulfide (D414C and V969C), Locked-1 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, Spike glycoprotein | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-10-19 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H13
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x2 Disulfide (G400C and V969C), Closed Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-10-19 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H16
 
 | | Structure of SARS-CoV-1 Spike Protein (S/native) at pH 5.5, Open Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-11-09 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.35534 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H11
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Closed Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-11-09 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H0Z
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Locked-122 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-11-09 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H0Y
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Locked-112 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, LINOLEIC ACID, ... | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-11-09 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H12
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x2 Disulfide (G400C and V969C), Locked-2 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-11-09 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.44681 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H0X
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Locked-1 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, LINOLEIC ACID, ... | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-11-09 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
6LRM
 
 | | Crystal structure of PDE4D catalytic domain in complex with arctigenin | | Descriptor: | 1,2-ETHANEDIOL, Arctigenin, MAGNESIUM ION, ... | | Authors: | Zhang, X.L, Li, M.J, Xu, Y.C. | | Deposit date: | 2020-01-16 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Identification of phosphodiesterase-4 as the therapeutic target of arctigenin in alleviating psoriatic skin inflammation.
J Adv Res, 33, 2021
|
|
