6KGR
 
 | | LSD1-FCPA-MPE N5 adduct model | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-[4-[5-fluoranyl-2-(trifluoromethyl)phenyl]phenyl]propanal, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Niwa, H, Sato, S, Handa, N, Umehara, T. | | Deposit date: | 2019-07-12 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Development and Structural Evaluation of N-Alkylated trans-2-Phenylcyclopropylamine-Based LSD1 Inhibitors.
Chemmedchem, 15, 2020
|
|
6K0F
 
 | | The crystal structure of cyclopenin-bound AsqJ quinary complex | | Descriptor: | (3~{R},3'~{S})-4-methyl-3'-phenyl-spiro[1~{H}-1,4-benzodiazepine-3,2'-oxirane]-2,5-dione, 2-OXOGLUTARIC ACID, FE (III) ION, ... | | Authors: | Liao, H.J, Chan, N.L. | | Deposit date: | 2019-05-06 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.634 Å) | | Cite: | The crystal structure of cyclopenin-bound AsqJ quinary complex
J.Am.Chem.Soc., 2020
|
|
6KGQ
 
 | | LSD1-FCPA-MPE five-membered ring adduct model | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-[4-[5-fluoranyl-2-(trifluoromethyl)phenyl]phenyl]propanal, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Niwa, H, Sato, S, Handa, N, Umehara, T. | | Deposit date: | 2019-07-12 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Development and Structural Evaluation of N-Alkylated trans-2-Phenylcyclopropylamine-Based LSD1 Inhibitors.
Chemmedchem, 15, 2020
|
|
6L8Z
 
 | | Crystal structure of ugt transferase mutant in complex with UPG | | Descriptor: | Glycosyltransferase, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Li, J, Shan, N, Yang, J.G, Liu, W.D, Sun, Y.X. | | Deposit date: | 2019-11-07 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Efficient O-Glycosylation of Triterpenes Enabled by Protein Engineering of Plant Glycosyltransferase UGT74AC1
Acs Catalysis, 2020
|
|
4UA2
 
 | | Crystal structure of dual function transcriptional regulator MerR from Bacillus megaterium MB1 | | Descriptor: | Regulatory protein | | Authors: | Lin, L.Y, Chang, C.C, Zou, X.W, Huang, C.C, Chan, N.L. | | Deposit date: | 2014-08-07 | | Release date: | 2015-07-22 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural basis of the mercury(II)-mediated conformational switching of the dual-function transcriptional regulator MerR
Nucleic Acids Res., 43, 2015
|
|
4UA1
 
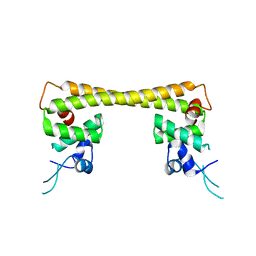 | | Crystal structure of dual function transcriptional regulator MerR form Bacillus megaterium MB1 in complex with mercury (II) ion | | Descriptor: | MERCURY (II) ION, Regulatory protein | | Authors: | Chang, C.C, Lin, L.Y, Zou, X.W, Huang, C.C, Chan, N.L. | | Deposit date: | 2014-08-07 | | Release date: | 2015-07-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structural basis of the mercury(II)-mediated conformational switching of the dual-function transcriptional regulator MerR
Nucleic Acids Res., 43, 2015
|
|
6LK0
 
 | | Crystal structure of human wild type TRIP13 | | Descriptor: | Pachytene checkpoint protein 2 homolog | | Authors: | Wang, Y, Huang, J, Li, B, Xue, H, Tricot, G, Hu, L, Xu, Z, Sun, X, Chang, S, Gao, L, Tao, Y, Xu, H, Xie, Y, Xiao, W, Yu, D, Kong, Y, Chen, G, Sun, X, Lian, F, Zhang, N, Wu, X, Mao, Z, Zhan, F, Zhu, W, Shi, J. | | Deposit date: | 2019-12-17 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Small-Molecule Inhibitor Targeting TRIP13 Suppresses Multiple Myeloma Progression.
Cancer Res., 80, 2020
|
|
3W28
 
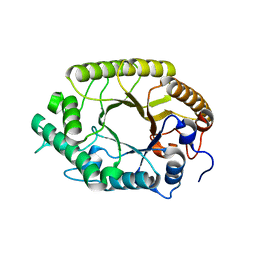 | | The high-resolution crystal structure of TsXylA, intracellular xylanase from /Thermoanaerobacterium saccharolyticum JW/SL-YS485/: the complex of the E251A mutant with xylotriose | | Descriptor: | Glycoside hydrolase family 10, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-alpha-D-xylopyranose | | Authors: | Han, X, Gao, J, Shang, N, Huang, C.-H, Ko, T.-P, Zhu, Z, Wiegel, J, Shao, W, Guo, R.-T. | | Deposit date: | 2012-11-27 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural and functional analyses of catalytic domain of GH10 xylanase from Thermoanaerobacterium saccharolyticum JW/SL-YS485
Proteins, 81, 2013
|
|
3W29
 
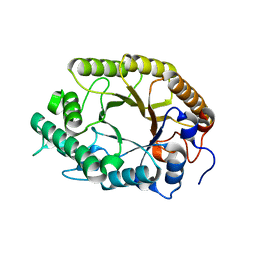 | | The high-resolution crystal structure of TsXylA, intracellular xylanase from /Thermoanaerobacterium saccharolyticum JW/SL-YS485/: the complex of the E251A mutant with xylotetraose | | Descriptor: | Glycoside hydrolase family 10, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-alpha-D-xylopyranose | | Authors: | Han, X, Gao, J, Shang, N, Huang, C.-H, Ko, T.-P, Zhu, Z, Wiegel, J, Shao, W, Guo, R.-T. | | Deposit date: | 2012-11-27 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural and functional analyses of catalytic domain of GH10 xylanase from Thermoanaerobacterium saccharolyticum JW/SL-YS485
Proteins, 81, 2013
|
|
3W27
 
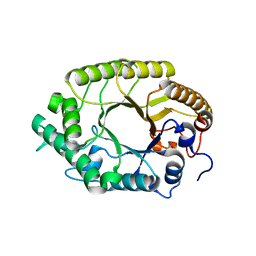 | | The high-resolution crystal structure of TsXylA, intracellular xylanase from /Thermoanaerobacterium saccharolyticum JW/SL-YS485/: the complex of the E251A mutant with xylobiose | | Descriptor: | Glycoside hydrolase family 10, beta-D-xylopyranose-(1-4)-alpha-D-xylopyranose | | Authors: | Han, X, Gao, J, Shang, N, Huang, C.-H, Ko, T.-P, Zhu, Z, Wiegel, J, Shao, W, Guo, R.-T. | | Deposit date: | 2012-11-27 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structural and functional analyses of catalytic domain of GH10 xylanase from Thermoanaerobacterium saccharolyticum JW/SL-YS485
Proteins, 81, 2013
|
|
3W25
 
 | | The high-resolution crystal structure of TsXylA, intracellular xylanase from /Thermoanaerobacterium saccharolyticum JW/SL-YS485/: the complex of the E146A mutant with xylobiose | | Descriptor: | Glycoside hydrolase family 10, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Han, X, Gao, J, Shang, N, Huang, C.-H, Ko, T.-P, Zhu, Z, Wiegel, J, Shao, W, Guo, R.-T. | | Deposit date: | 2012-11-27 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structural and functional analyses of catalytic domain of GH10 xylanase from Thermoanaerobacterium saccharolyticum JW/SL-YS485
Proteins, 81, 2013
|
|
3W26
 
 | | The high-resolution crystal structure of TsXylA, intracellular xylanase from /Thermoanaerobacterium saccharolyticum JW/SL-YS485/: the complex of the E146A mutant with xylotriose | | Descriptor: | Glycoside hydrolase family 10, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Han, X, Gao, J, Shang, N, Huang, C.-H, Ko, T.-P, Zhu, Z, Wiegel, J, Shao, W, Guo, R.-T. | | Deposit date: | 2012-11-27 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and functional analyses of catalytic domain of GH10 xylanase from Thermoanaerobacterium saccharolyticum JW/SL-YS485
Proteins, 81, 2013
|
|
3W24
 
 | | The high-resolution crystal structure of TsXylA, intracellular xylanase from Thermoanaerobacterium saccharolyticum JW/SL-YS485 | | Descriptor: | Glycoside hydrolase family 10 | | Authors: | Han, X, Gao, J, Shang, N, Huang, C.-H, Ko, T.-P, Zhu, Z, Wiegel, J, Shao, W, Guo, R.-T. | | Deposit date: | 2012-11-27 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural and functional analyses of catalytic domain of GH10 xylanase from Thermoanaerobacterium saccharolyticum JW/SL-YS485
Proteins, 81, 2013
|
|
7CLF
 
 | | PigF with SAH | | Descriptor: | ACETATE ION, Methyltransferase domain-containing protein, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Qiu, S, Xu, D, Han, N, Sun, B, Ran, T, Wang, W. | | Deposit date: | 2020-07-20 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.982 Å) | | Cite: | Crystal structures of PigF, an O-methyltransferase involved in the prodigiosin synthetic pathway, reveal an induced-fit substrate-recognition mechanism.
Iucrj, 9, 2022
|
|
7CLU
 
 | | PigF with SAH | | Descriptor: | ACETATE ION, GLYCEROL, Methyltransferase domain-containing protein | | Authors: | Qiu, S, Xu, D, Han, N, Sun, B, Ran, T, Wang, W. | | Deposit date: | 2020-07-22 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of PigF, an O-methyltransferase involved in the prodigiosin synthetic pathway, reveal an induced-fit substrate-recognition mechanism.
Iucrj, 9, 2022
|
|
3UUX
 
 | | Crystal structure of yeast Fis1 in complex with Mdv1 fragment containing N-terminal extension and coiled coil domains | | Descriptor: | Mitochondria fission 1 protein, Mitochondrial division protein 1 | | Authors: | Zhang, Y, Chan, N.C, Gristick, H, Chan, D.C. | | Deposit date: | 2011-11-28 | | Release date: | 2012-02-08 | | Last modified: | 2012-04-25 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Crystal structure of mitochondrial fission complex reveals scaffolding function for mitochondrial division 1 (mdv1) coiled coil.
J.Biol.Chem., 287, 2012
|
|
6SES
 
 | | Tubulin-B2 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Guo, B, Rodriguez-Gabin, A, Prota, A.E, Muehlethaler, T, Zhang, N, Ye, K, Steinmetz, M.O, Band Horwitz, S, Smith III, A.B, McDaid, H.M. | | Deposit date: | 2019-07-30 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Refinement of the Tubulin Ligand (+)-Discodermolide to Attenuate Chemotherapy-Mediated Senescence.
Mol.Pharmacol., 98, 2020
|
|
6M7D
 
 | | Structure of ncleoprotein of sendai virus | | Descriptor: | Nucleoprotein, RNA (5'-R(P*UP*UP*UP*UP*UP*U)-3') | | Authors: | Shen, Q, Shan, H, Zhang, N. | | Deposit date: | 2020-03-18 | | Release date: | 2021-03-24 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | structure of the nucleocapsid of sendai virus at 2.9 Angstroms resolution
To Be Published
|
|
3L6V
 
 | | Crystal Structure of the Xanthomonas campestris Gyrase A C-terminal Domain | | Descriptor: | DNA gyrase subunit A | | Authors: | Hsieh, T.J, Yen, T.J, Lin, T.S, Chang, H.T, Huang, S.Y, Farh, L, Chan, N.L. | | Deposit date: | 2009-12-26 | | Release date: | 2010-03-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Twisting of the DNA binding surface by a beta-strand-bearing proline modulates DNA gyrase activity
To be Published
|
|
7E38
 
 | | Crystal structure of deoxypodophyllotoxin synthase from Sinopodophyllum hexandrum in complex with yatein and succinate | | Descriptor: | (3~{R},4~{R})-4-(1,3-benzodioxol-5-ylmethyl)-3-[(3,4,5-trimethoxyphenyl)methyl]oxolan-2-one, (3~{S},4~{S})-4-(1,3-benzodioxol-5-ylmethyl)-3-[(3,4,5-trimethoxyphenyl)methyl]oxolan-2-one, Deoxypodophyllotoxin synthase, ... | | Authors: | Wu, M.-H, Chang, W.-c, Chien, T.-C, Chan, N.-L. | | Deposit date: | 2021-02-08 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Mechanistic analysis of carbon-carbon bond formation by deoxypodophyllotoxin synthase.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7E37
 
 | | Crystal structure of deoxypodophyllotoxin synthase from Sinopodophyllum hexandrum in complex with 2-oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, Deoxypodophyllotoxin synthase, FE (III) ION | | Authors: | Wu, M.-H, Lin, H.-Y, Chang, W.-c, Chien, T.-C, Chan, N.-L. | | Deposit date: | 2021-02-08 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Mechanistic analysis of carbon-carbon bond formation by deoxypodophyllotoxin synthase.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7C1L
 
 | | Crystal structure of the starter condensation domain of rhizomide synthetase RzmA mutant R148A in complex with C8-CoA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Non-ribosomal peptide synthetase modules, OCTANOYL-COENZYME A | | Authors: | Zhong, L, Diao, X, Zhang, N, Li, F.W, Zhou, H.B, Chen, H.N, Ren, X, Zhang, Y, Wu, D, Bian, X. | | Deposit date: | 2020-05-05 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Engineering and elucidation of the lipoinitiation process in nonribosomal peptide biosynthesis.
Nat Commun, 12, 2021
|
|
7C1U
 
 | | Crystal structure of the starter condensation domain of rhizomide synthetase RzmA mutant H140V/R148A in a "product-released" conformation | | Descriptor: | Non-ribosomal peptide synthetase modules | | Authors: | Zhong, L, Diao, X, Zhang, N, Li, F.W, Zhou, H.B, Chen, H.N, Ren, X, Zhang, Y, Wu, D, Bian, X. | | Deposit date: | 2020-05-05 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Engineering and elucidation of the lipoinitiation process in nonribosomal peptide biosynthesis.
Nat Commun, 12, 2021
|
|
7C1H
 
 | | Crystal structure of the starter condensation domain of rhizomide synthetase RzmA | | Descriptor: | Non-ribosomal peptide synthetase modules | | Authors: | Zhong, L, Diao, X, Zhang, N, Li, F.W, Zhou, H.B, Chen, H.N, Ren, X, Zhang, Y, Wu, D, Bian, X. | | Deposit date: | 2020-05-04 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Engineering and elucidation of the lipoinitiation process in nonribosomal peptide biosynthesis.
Nat Commun, 12, 2021
|
|
7C1R
 
 | | Crystal structure of the starter condensation domain of rhizomide synthetase RzmA mutant H140A/R148A in complex with C8-CoA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Non-ribosomal peptide synthetase modules, OCTANOYL-COENZYME A | | Authors: | Zhong, L, Diao, X, Zhang, N, Li, F.W, Zhou, H.B, Chen, H.N, Ren, X, Zhang, Y, Wu, D, Bian, X. | | Deposit date: | 2020-05-05 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Engineering and elucidation of the lipoinitiation process in nonribosomal peptide biosynthesis.
Nat Commun, 12, 2021
|
|
