4YY8
 
 | | Crystal Structure Analysis of Kelch protein from Plasmodium falciparum | | Descriptor: | GLYCEROL, Kelch protein, UNKNOWN ATOM OR ION | | Authors: | Jiang, D.Q, Tempel, W, Loppnau, P, Graslund, S, He, H, Ravichandran, M, Seitova, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, El Bakkouri, M, Senisterra, G, Osman, K.T, Lovato, D.V, Hui, R, Hutchinson, A, Lin, Y.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-03-23 | | Release date: | 2015-04-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal Structure Analysis of Kelch protein from Plasmodium falciparum.
to be published
|
|
2FQD
 
 | | Crystal Structures of E. coli Laccase CueO under different copper binding situations | | Descriptor: | Blue copper oxidase cueO, CITRIC ACID, COPPER (II) ION, ... | | Authors: | Li, X, Wei, Z, Zhang, M, Teng, M, Gong, W. | | Deposit date: | 2006-01-18 | | Release date: | 2007-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of E. coli laccase CueO at different copper concentrations.
Biochem.Biophys.Res.Commun., 354, 2007
|
|
5T89
 
 | | Crystal structure of VEGF-A in complex with VEGFR-1 domains D1-6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Vascular endothelial growth factor A, ... | | Authors: | Markovic-Mueller, S, Stuttfeld, E, Asthana, M, Weinert, T, Bliven, S, Goldie, K.N, Kisko, K, Capitani, G, Ballmer-Hofer, K. | | Deposit date: | 2016-09-07 | | Release date: | 2017-02-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structure of the Full-length VEGFR-1 Extracellular Domain in Complex with VEGF-A.
Structure, 25, 2017
|
|
2FQF
 
 | | Crystal Structures of E. coli Laccase CueO under different copper binding situations | | Descriptor: | Blue copper oxidase cueO, CITRIC ACID, COPPER (II) ION, ... | | Authors: | Li, X, Wei, Z, Zhang, M, Teng, M, Gong, W. | | Deposit date: | 2006-01-18 | | Release date: | 2007-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of E. coli laccase CueO at different copper concentrations.
Biochem.Biophys.Res.Commun., 354, 2007
|
|
5FYW
 
 | | Transcription initiation complex structures elucidate DNA opening (OC) | | Descriptor: | DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB1, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB11, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB2, ... | | Authors: | Plaschka, C, Hantsche, M, Dienemann, C, Burzinski, C, Plitzko, J, Cramer, P. | | Deposit date: | 2016-03-10 | | Release date: | 2016-05-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.35 Å) | | Cite: | Transcription Initiation Complex Structures Elucidate DNA Opening
Nature, 533, 2016
|
|
2FQE
 
 | | Crystal Structures of E. coli Laccase CueO under different copper binding situations | | Descriptor: | Blue copper oxidase cueO, CITRIC ACID, COPPER (II) ION, ... | | Authors: | Li, X, Wei, Z, Zhang, M, Teng, M, Gong, W. | | Deposit date: | 2006-01-18 | | Release date: | 2007-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structures of E. coli laccase CueO at different copper concentrations.
Biochem.Biophys.Res.Commun., 354, 2007
|
|
2FQG
 
 | | Crystal Structures of E. coli Laccase CueO under different copper binding situations | | Descriptor: | Blue copper oxidase cueO, CITRIC ACID, COPPER (II) ION, ... | | Authors: | Li, X, Wei, Z, Zhang, M, Teng, M, Gong, W. | | Deposit date: | 2006-01-18 | | Release date: | 2007-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of E. coli laccase CueO at different copper concentrations.
Biochem.Biophys.Res.Commun., 354, 2007
|
|
5AGS
 
 | | Crystal structure of the LeuRS editing domain of Mycobacterium tuberculosis in complex with the adduct 3-(Aminomethyl)-4-bromo-7-ethoxybenzo[c][1,2]oxaborol-1(3H)-ol-AMP | | Descriptor: | 3-(AMINOMETHYL)-4-BROMO-7-ETHOXYBENZO[C][1,2]OXABOROL-1(3H)-OL-MODIFIED ADENOSINE, LEUCYL-TRNA SYNTHETASE, METHIONINE | | Authors: | Palencia, A, Li, X, Alley, M.R.K, Ding, C, Easom, E.E, Hernandez, V, Meewan, M, Mohan, M, Rock, F.L, Franzblau, S.G, Wang, Y, Lenaerts, A.J, Parish, T, Cooper, C.B, Waters, M.G, Ma, Z, Mendoza, A, Barros, D, Cusack, S, Plattner, J.J. | | Deposit date: | 2015-02-03 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Discovery of Novel Oral Protein Synthesis Inhibitors of Mycobacterium Tuberculosis that Target Leucyl-tRNA Synthetase.
Antimicrob.Agents Chemother., 60, 2016
|
|
6LXB
 
 | | X-ray structure of human PPARalpha ligand binding domain-saroglitazar co-crystals obtained by soaking | | Descriptor: | (2S)-2-ethoxy-3-[4-[2-[2-methyl-5-(4-methylsulfanylphenyl)pyrrol-1-yl]ethoxy]phenyl]propanoic acid, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Honda, A, Ishikawa, R, Akahane, M, Oyama, T, Ishii, I. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
3PZD
 
 | | Structure of the myosin X MyTH4-FERM/DCC complex | | Descriptor: | GLYCEROL, Myosin-X, Netrin receptor DCC | | Authors: | Wei, Z, Yan, J, Pan, L, Zhang, M. | | Deposit date: | 2010-12-14 | | Release date: | 2011-02-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Cargo recognition mechanism of myosin X revealed by the structure of its tail MyTH4-FERM tandem in complex with the DCC P3 domain
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
8C2D
 
 | | 14-3-3 in complex with Pyrin pS208 | | Descriptor: | 14-3-3 protein sigma, MAGNESIUM ION, Pyrin pS208 peptide | | Authors: | Lau, R, Ottmann, C, Hann, M. | | Deposit date: | 2022-12-22 | | Release date: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure and ligandability of the 14-3-3/pyrin interface.
Biochem.Biophys.Res.Commun., 651, 2023
|
|
5FZ5
 
 | | Transcription initiation complex structures elucidate DNA opening (CC) | | Descriptor: | DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB1, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB11, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB2, ... | | Authors: | Plaschka, C, Hantsche, M, Dienemann, C, Burzinski, C, Plitzko, J, Cramer, P. | | Deposit date: | 2016-03-10 | | Release date: | 2016-05-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.8 Å) | | Cite: | Transcription Initiation Complex Structures Elucidate DNA Opening
Nature, 533, 2016
|
|
6AVJ
 
 | | Crystal structure of human Mitochondrial inner NEET protein (MiNT)/CISD3 | | Descriptor: | CDGSH iron-sulfur domain-containing protein 3, mitochondrial, FE2/S2 (INORGANIC) CLUSTER | | Authors: | Lipper, C.H, Karmi, O, Sohn, Y.S, Darash-Yahana, M, Lammert, H, Song, L, Liu, A, Mittler, R, Nechushtai, R, Onuchic, J.N, Jennings, P.A. | | Deposit date: | 2017-09-02 | | Release date: | 2017-12-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the human monomeric NEET protein MiNT and its role in regulating iron and reactive oxygen species in cancer cells.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2PLW
 
 | | Crystal structure of a ribosomal RNA methyltransferase, putative, from Plasmodium falciparum (PF13_0052). | | Descriptor: | Ribosomal RNA methyltransferase, putative, S-ADENOSYLMETHIONINE, ... | | Authors: | Wernimont, A.K, Hassanali, A, Lin, L, Lew, J, Zhao, Y, Ravichandran, M, Wasney, G, Vedadi, M, Kozieradzki, I, Schapira, M, Bochkarev, A, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Hui, R, Qiu, W, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-04-20 | | Release date: | 2007-05-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a ribosomal RNA methyltransferase, putative, from Plasmodium falciparum (PF13_0052).
To be Published
|
|
7YFM
 
 | | Structure of GluN1b-GluN2D NMDA receptor in complex with agonists glycine and glutamate. | | Descriptor: | Glutamate receptor ionotropic, NMDA 2D, Isoform 6 of Glutamate receptor ionotropic, ... | | Authors: | Zhang, J.L, Zhu, S.J, Zhang, M. | | Deposit date: | 2022-07-08 | | Release date: | 2023-03-29 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8C28
 
 | | 14-3-3 in complex with PyrinpS208pS242 | | Descriptor: | 14-3-3 protein sigma, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Pyrin | | Authors: | Lau, R, Ottmann, C, Hann, M. | | Deposit date: | 2022-12-21 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 14-3-3/Pyrin complex
To Be Published
|
|
7YFF
 
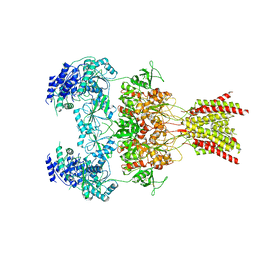 | | Structure of GluN1a-GluN2D NMDA receptor in complex with agonist glycine and competitive antagonist CPP. | | Descriptor: | (2R)-4-(3-phosphonopropyl)piperazine-2-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J.L, Zhu, S.J, Zhang, M. | | Deposit date: | 2022-07-08 | | Release date: | 2023-04-12 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7YFL
 
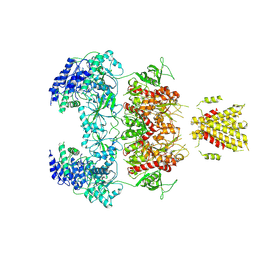 | | Structure of GluN1a-GluN2D NMDA receptor in complex with agonists glycine and glutamate. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Zhang, J.L, Zhu, S.J, Zhang, M. | | Deposit date: | 2022-07-08 | | Release date: | 2023-04-12 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7YFO
 
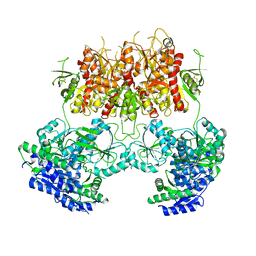 | |
7YFR
 
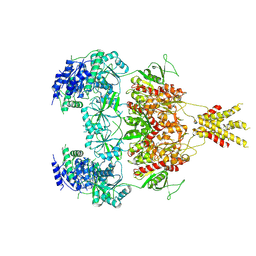 | | Structure of GluN1a E698C-GluN2D NMDA receptor in cystines non-crosslinked state. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Zhang, J.L, Zhu, S.J, Zhang, M. | | Deposit date: | 2022-07-09 | | Release date: | 2023-04-12 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8C2Y
 
 | |
5B8D
 
 | | Crystal structure of a low occupancy fragment candidate (N-(4-Methyl-1,3-thiazol-2-yl)propanamide) bound adjacent to the ubiquitin binding pocket of the HDAC6 zinc-finger domain | | Descriptor: | FORMIC ACID, Histone deacetylase 6, SODIUM ION, ... | | Authors: | Harding, R.J, Tempel, W, Collins, P, Pearce, N, Brandao-Neto, J, Douangamath, A, Ravichandran, M, Schapira, M, Bountra, C, Edwards, A.M, von Delft, F, Santhakumar, V, Arrowsmith, C.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-14 | | Release date: | 2016-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Small Molecule Antagonists of the Interaction between the Histone Deacetylase 6 Zinc-Finger Domain and Ubiquitin.
J. Med. Chem., 60, 2017
|
|
5Z0Y
 
 | |
6LXC
 
 | | X-ray structure of human PPARalpha ligand binding domain-saroglitazar co-crystals obtained by delipidation and cross-seeding | | Descriptor: | (2S)-2-ethoxy-3-[4-[2-[2-methyl-5-(4-methylsulfanylphenyl)pyrrol-1-yl]ethoxy]phenyl]propanoic acid, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Honda, A, Ishikawa, R, Akahane, M, Oyama, T, Ishii, I. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
7CDB
 
 | | Structure of GABARAPL1 in complex with GABA(A) receptor gamma 2 | | Descriptor: | ACETATE ION, CITRIC ACID, Gamma-aminobutyric acid receptor subunit gamma-2, ... | | Authors: | Li, J, Ye, J, Zhu, R, Kong, C, Zhang, M, Wang, C. | | Deposit date: | 2020-06-19 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Structural basis of GABARAP-mediated GABA A receptor trafficking and functions on GABAergic synaptic transmission.
Nat Commun, 12, 2021
|
|
