6J6F
 
 | | Ligand binding domain 1 and 2 of Talaromyces marneffei Mp1 protein | | Descriptor: | Envelope glycoprotein, NICKEL (II) ION | | Authors: | Lam, W.H, Zhang, H, Hao, Q. | | Deposit date: | 2019-01-15 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Talaromyces marneffeiMp1 Protein, a Novel Virulence Factor, Carries Two Arachidonic Acid-Binding Domains To Suppress Inflammatory Responses in Hosts.
Infect. Immun., 87, 2019
|
|
3R1B
 
 | | Open crystal structure of cytochrome P450 2B4 covalently bound to the mechanism-based inactivator tert-butylphenylacetylene | | Descriptor: | (4-tert-butylphenyl)acetaldehyde, 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, Cytochrome P450 2B4, ... | | Authors: | Gay, S.C, Zhang, H, Stout, C.D, Hollenberg, P.F, Halpert, J.R. | | Deposit date: | 2011-03-09 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Analysis of Mammalian Cytochrome P450 2B4 Covalently Bound to the Mechanism-Based Inactivator tert-Butylphenylacetylene: Insight into Partial Enzymatic Activity.
Biochemistry, 50, 2011
|
|
4B4H
 
 | | Thermobifida fusca cellobiohydrolase Cel6B(E3) catalytic domain | | Descriptor: | BETA-1,4-EXOCELLULASE | | Authors: | Sandgren, M, Wu, M, Stahlberg, J, Karkehabadi, S, Mitchinson, C, Kelemen, B.R, Larenas, E.A, Hansson, H. | | Deposit date: | 2012-07-30 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Structure of a Bacterial Cellobiohydrolase: The Catalytic Core of the Thermobifida Fusca Family Gh6 Cellobiohydrolase Cel6B.
J.Mol.Biol., 425, 2013
|
|
4CSI
 
 | | Crystal structure of the thermostable Cellobiohydrolase Cel7A from the fungus Humicola grisea var. thermoidea. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CELLULASE, DI(HYDROXYETHYL)ETHER | | Authors: | Haddad-Momeni, M, Goedegebuur, F, Hansson, H, Karkehabadi, S, Askarieh, G, Mitchinson, C, Larenas, E, Stahlberg, J, Sandgren, M. | | Deposit date: | 2014-03-07 | | Release date: | 2014-09-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Expression, Crystal Structure and Cellulase Activity of the Thermostable Cellobiohydrolase Cel7A from the Fungus Humicola Grisea Var. Thermoidea.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5X1W
 
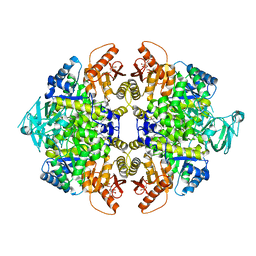 | | PKM2 in complex with compound 5 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 4-[2,3-bis(chloranyl)phenyl]carbonyl-N-[2-[[4-[2,3-bis(chloranyl)phenyl]carbonyl-1-methyl-pyrrol-2-yl]carbonylamino]ethyl]-1-methyl-pyrrole-2-carboxamide, Pyruvate kinase PKM | | Authors: | Matsui, Y, Hanzawa, H. | | Deposit date: | 2017-01-27 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery and structure-guided fragment-linking of 4-(2,3-dichlorobenzoyl)-1-methyl-pyrrole-2-carboxamide as a pyruvate kinase M2 activator
Bioorg. Med. Chem., 25, 2017
|
|
4B4F
 
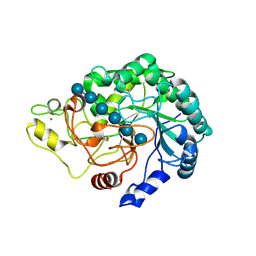 | | Thermobifida fusca Cel6B(E3) co-crystallized with cellobiose | | Descriptor: | BETA-1,4-EXOCELLULASE, CALCIUM ION, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Sandgren, M, Wu, M, Stahlberg, J, Karkehabadi, S, Mitchinson, C, Kelemen, B.R, Larenas, E.A, Hansson, H. | | Deposit date: | 2012-07-30 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structure of a Bacterial Cellobiohydrolase: The Catalytic Core of the Thermobifida Fusca Family Gh6 Cellobiohydrolase Cel6B.
J.Mol.Biol., 425, 2013
|
|
5ZYL
 
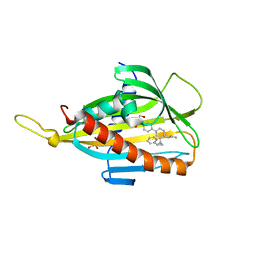 | | Crystal structure of CERT START domain in complex with compound E25A | | Descriptor: | 2-[4-[4-cyclopentyl-3-[(1~{S},2~{R})-2-pyridin-2-ylcyclopropyl]phenyl]phenyl]sulfonylethanol, GLYCEROL, LIPID-TRANSFER PROTEIN CERT | | Authors: | Suzuki, M, Nakao, N, Ueno, M, Sakai, S, Egawa, D, Hanzawa, H, Kawasaki, S, Kumagai, K, Kobayashi, S, Hanada, K. | | Deposit date: | 2018-05-25 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Natural ligand-nonmimetic inhibitors of the lipid-transfer protein CERT
Commun Chem, 2019
|
|
5ZYI
 
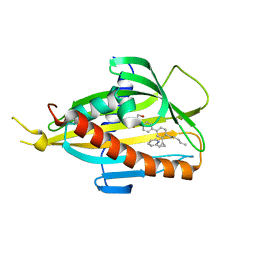 | | Crystal structure of CERT START domain in complex with compound E16 | | Descriptor: | 2-[4-[4-pentyl-3-[(1~{S},2~{R})-2-pyridin-2-ylcyclopropyl]phenyl]phenyl]sulfonylethanol, LIPID-TRANSFER PROTEIN CERT | | Authors: | Suzuki, M, Nakao, N, Ueno, M, Sakai, S, Egawa, D, Hanzawa, H, Kawasaki, S, Kumagai, K, Kobayashi, S, Hanada, K. | | Deposit date: | 2018-05-25 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Natural ligand-nonmimetic inhibitors of the lipid-transfer protein CERT
Commun Chem, 2019
|
|
5ZYJ
 
 | | Crystal structure of CERT START domain in complex with compound E16A | | Descriptor: | 2-[4-[4-pentyl-3-[(1~{S},2~{R})-2-pyridin-2-ylcyclopropyl]phenyl]phenyl]sulfonylethanol, GLYCEROL, LIPID-TRANSFER PROTEIN CERT | | Authors: | Suzuki, M, Nakao, N, Ueno, M, Sakai, S, Egawa, D, Hanzawa, H, Kawasaki, S, Kumagai, K, Kobayashi, S, Hanada, K. | | Deposit date: | 2018-05-25 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Natural ligand-nonmimetic inhibitors of the lipid-transfer protein CERT
Commun Chem, 2019
|
|
7V6D
 
 | | Structure of lipase B from Lasiodiplodia theobromae | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Lipase B | | Authors: | Xue, B, Zhang, H.F, Nguyen, G.K.T, Yew, W.S. | | Deposit date: | 2021-08-20 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Novel Lipase from Lasiodiplodia theobromae Efficiently Hydrolyses C8-C10 Methyl Esters for the Preparation of Medium-Chain Triglycerides' Precursors.
Int J Mol Sci, 22, 2021
|
|
7F61
 
 | |
1B45
 
 | | ALPHA-CNIA CONOTOXIN FROM CONUS CONSORS, NMR, 43 STRUCTURES | | Descriptor: | ALPHA-CNIA | | Authors: | Favreau, P, Krimm, I, Le Gall, F, Bobenrieth, M.J, Lamthanh, H, Bouet, F, Servent, D, Molgo, J, Menez, A, Letourneux, Y, Lancelin, J.M. | | Deposit date: | 1999-01-05 | | Release date: | 1999-07-09 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Biochemical characterization and nuclear magnetic resonance structure of novel alpha-conotoxins isolated from the venom of Conus consors.
Biochemistry, 38, 1999
|
|
3T88
 
 | | Crystal structure of Escherichia coli MenB in complex with substrate analogue, OSB-NCoA | | Descriptor: | 1,4-Dihydroxy-2-naphthoyl-CoA synthase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Li, H.-J, Li, X, Liu, N, Zhang, H, Truglio, J, Mishra, S, Kisker, C, Garcia-Diaz, M, Tonge, P. | | Deposit date: | 2011-08-01 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Mechanism of the Intramolecular Claisen Condensation Reaction Catalyzed by MenB, a Crotonase Superfamily Member.
Biochemistry, 50, 2011
|
|
7XW9
 
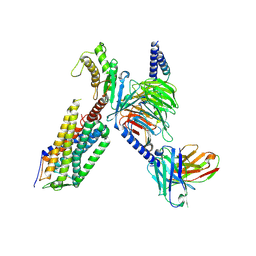 | | Cryo-EM structure of the TRH-bound human TRHR-Gq complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(q) subunit alpha, ... | | Authors: | Ji, S, Dong, Y, Chen, L, Zang, S, Shen, D, Guo, J, Qin, J, Zhang, H, Wang, W, Shen, Q, Mao, C, Zhang, Y. | | Deposit date: | 2022-05-26 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Molecular basis for the activation of thyrotropin-releasing hormone receptor.
Cell Discov, 8, 2022
|
|
7XOU
 
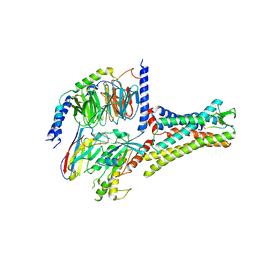 | | Structural insights into human brain gut peptide cholecystokinin receptors | | Descriptor: | CCK-8, Cholecystokinin receptor type A, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Ding, Y, Zhang, H, Liao, Y, Chen, L, Ji, S. | | Deposit date: | 2022-05-01 | | Release date: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into human brain-gut peptide cholecystokinin receptors.
Cell Discov, 8, 2022
|
|
7XOW
 
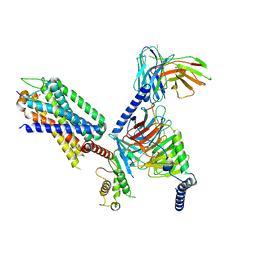 | | Structural insights into human brain gut peptide cholecystokinin receptors | | Descriptor: | Gastrin, Gastrin/cholecystokinin type B receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Ding, Y, Zhang, H, Liao, Y, Chen, L, Ji, S. | | Deposit date: | 2022-05-01 | | Release date: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into human brain-gut peptide cholecystokinin receptors.
Cell Discov, 8, 2022
|
|
7XOV
 
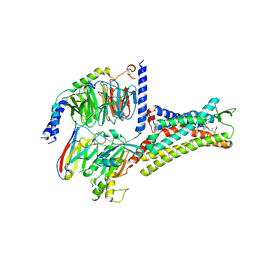 | | Structural insights into human brain gut peptide cholecystokinin receptors | | Descriptor: | 2-[2-[[4-(4-chloranyl-2,5-dimethoxy-phenyl)-5-(2-cyclohexylethyl)-1,3-thiazol-2-yl]carbamoyl]-5,7-dimethyl-indol-1-yl]ethanoic acid, Cholecystokinin receptor type A, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Ding, Y, Zhang, H, Liao, Y, Chen, L, Ji, S. | | Deposit date: | 2022-05-01 | | Release date: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into human brain-gut peptide cholecystokinin receptors.
Cell Discov, 8, 2022
|
|
3T89
 
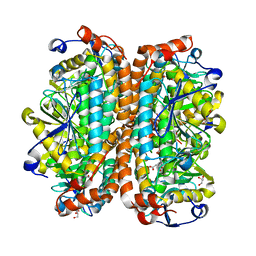 | | Crystal structure of Escherichia coli MenB, the 1,4-dihydroxy-2-naphthoyl-CoA synthase in vitamin K2 biosynthesis | | Descriptor: | 1,4-Dihydroxy-2-naphthoyl-CoA synthase, GLYCEROL, MALONATE ION | | Authors: | Li, H.-J, Li, X, Liu, N, Zhang, H, Truglio, J, Mishra, S, Kisker, C, Garcia-Diaz, M, Tonge, P. | | Deposit date: | 2011-08-01 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Mechanism of the Intramolecular Claisen Condensation Reaction Catalyzed by MenB, a Crotonase Superfamily Member.
Biochemistry, 50, 2011
|
|
3ZZ1
 
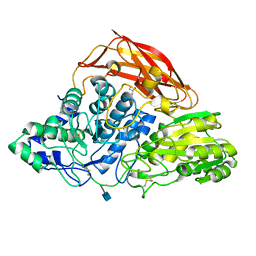 | | Crystal structure of a glycoside hydrolase family 3 beta-glucosidase, Bgl1 from Hypocrea jecorina at 2.1A resolution. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-D-GLUCOSIDE GLUCOHYDROLASE, GLYCEROL | | Authors: | Sandgren, M, Kaper, T, Mikkelsen, N.E, Hansson, H, Piens, K, Gudmundsson, M, Larenas, E, Kelemen, B, Karkehabadi, S. | | Deposit date: | 2011-08-31 | | Release date: | 2012-12-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biochemical Characterization and Crystal Structures of a Fungal Family 3 Beta-Glucosidase, Cel3A from Hypocrea Jecorina.
J.Biol.Chem., 289, 2014
|
|
3UAS
 
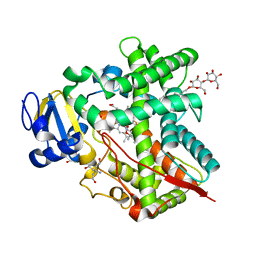 | | Cytochrome P450 2B4 covalently bound to the mechanism-based inactivator 9-ethynylphenanthrene | | Descriptor: | 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, Cytochrome P450 2B4, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Gay, S.C, Zhang, H, Shah, M.B, Stout, C.D, Halpert, J.R, Hollenberg, P.F. | | Deposit date: | 2011-10-21 | | Release date: | 2013-01-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.939 Å) | | Cite: | Potent Mechanism-Based Inactivation of Cytochrome P450 2B4 by 9-Ethynylphenanthrene: Implications for Allosteric Modulation of Cytochrome P450 Catalysis.
Biochemistry, 52, 2013
|
|
6AAX
 
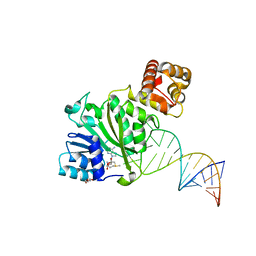 | | Crystal structure of TFB1M and h45 with SAM in homo sapiens | | Descriptor: | DI(HYDROXYETHYL)ETHER, Dimethyladenosine transferase 1, mitochondrial, ... | | Authors: | Liu, X, Shen, S, Wu, P, Li, F, Gong, Q, Wu, J, Zhang, H, Shi, Y. | | Deposit date: | 2018-07-19 | | Release date: | 2019-06-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.994 Å) | | Cite: | Structural insights into dimethylation of 12S rRNA by TFB1M: indispensable role in translation of mitochondrial genes and mitochondrial function.
Nucleic Acids Res., 47, 2019
|
|
5ZYH
 
 | | Crystal structure of CERT START domain in complex with compound E5 | | Descriptor: | 2-[4-[3-~{tert}-butyl-5-[(1~{R},2~{S})-2-pyridin-2-ylcyclopropyl]phenyl]phenyl]sulfonylethanol, LIPID-TRANSFER PROTEIN CERT | | Authors: | Suzuki, M, Nakao, N, Ueno, M, Sakai, S, Egawa, D, Hanzawa, H, Kawasaki, S, Kumagai, K, Kobayashi, S, Hanada, K. | | Deposit date: | 2018-05-25 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Natural ligand-nonmimetic inhibitors of the lipid-transfer protein CERT
Commun Chem, 2019
|
|
5ZYK
 
 | | Crystal structure of CERT START domain in complex with compound E25 | | Descriptor: | 2-[4-[4-cyclopentyl-3-[(1~{S},2~{R})-2-pyridin-2-ylcyclopropyl]phenyl]phenyl]sulfonylethanol, LIPID-TRANSFER PROTEIN CERT | | Authors: | Suzuki, M, Nakao, N, Ueno, M, Sakai, S, Egawa, D, Hanzawa, H, Kawasaki, S, Kumagai, K, Kobayashi, S, Hanada, K. | | Deposit date: | 2018-05-25 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Natural ligand-nonmimetic inhibitors of the lipid-transfer protein CERT
Commun Chem, 2019
|
|
5ZYG
 
 | | Crystal structure of CERT START domain in complex with compound B5 | | Descriptor: | 2-[4-[3-~{tert}-butyl-5-(2-pyridin-2-ylethyl)phenyl]phenyl]sulfonylethanol, LIPID-TRANSFER PROTEIN CERT | | Authors: | Suzuki, M, Nakao, N, Ueno, M, Sakai, S, Egawa, D, Hanzawa, H, Kawasaki, S, Kumagai, K, Kobayashi, S, Hanada, K. | | Deposit date: | 2018-05-25 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Natural ligand-nonmimetic inhibitors of the lipid-transfer protein CERT
Commun Chem, 2019
|
|
6LG4
 
 | |
