7OCB
 
 | | Crystal structure of Spindlin1 in complex with the inhibitor XY49-92B | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 7-[3-(1,3-dihydroisoindol-2-yl)propoxy]-2N-[2-(dimethylamino)ethyl]-6-methoxy-4N-(1-propan-2-ylpiperidin-4-yl)quinazoline-2,4-diamine, CHLORIDE ION, ... | | 著者 | Johansson, C, Krojer, T, Park, K, Xiong, Y, Jin, J, Oppermann, U. | | 登録日 | 2021-04-26 | | 公開日 | 2022-05-04 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.42 Å) | | 主引用文献 | Crystal structure of Spindlin1 in complex with the inhibitor XY49-92B
To Be Published
|
|
7BTA
 
 | |
7BTC
 
 | |
7BTD
 
 | |
6WIP
 
 | | Class A beta-lactamase from Micromonospora aurantiaca ATCC 27029 | | 分子名称: | ACETATE ION, Beta-lactamase, SULFATE ION | | 著者 | Chang, C, Tesar, C, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-04-10 | | 公開日 | 2020-04-22 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | class A beta-lactamase from
Micromonospora aurantiaca ATCC 27029
To Be Published
|
|
6WKP
 
 | | Crystal structure of RNA-binding domain of nucleocapsid phosphoprotein from SARS CoV-2, monoclinic crystal form | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Nucleoprotein, ZINC ION | | 著者 | Chang, C, Michalska, K, Jedrzejczak, R, Maltseva, N, Endres, M, Godzik, A, Kim, Y, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-04-16 | | 公開日 | 2020-04-29 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.67 Å) | | 主引用文献 | Epitopes recognition of SARS-CoV-2 nucleocapsid RNA binding domain by human monoclonal antibodies.
Iscience, 27, 2024
|
|
7BWQ
 
 | | Structure of nonstructural protein Nsp9 from SARS-CoV-2 | | 分子名称: | Nsp9, SULFATE ION | | 著者 | Zhang, C, Chen, Y, Li, L, Su, D. | | 登録日 | 2020-04-15 | | 公開日 | 2021-07-21 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.954 Å) | | 主引用文献 | Structural basis for the multimerization of nonstructural protein nsp9 from SARS-CoV-2.
Mol Biomed, 1, 2020
|
|
6X4I
 
 | | Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with 3'-uridinemonophosphate | | 分子名称: | 1,2-ETHANEDIOL, 3'-URIDINEMONOPHOSPHATE, SODIUM ION, ... | | 著者 | Chang, C, Kim, Y, Maltseva, N, Jedrzejczak, R, Endres, M, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-05-22 | | 公開日 | 2020-06-03 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Tipiracil binds to uridine site and inhibits Nsp15 endoribonuclease NendoU from SARS-CoV-2.
Commun Biol, 4, 2021
|
|
4WIW
 
 | | Crystal structure of C-terminal domain of putative chitinase from Desulfitobacterium hafniense DCB-2 | | 分子名称: | CALCIUM ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Chang, C, Tesar, C, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2014-09-26 | | 公開日 | 2014-10-08 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.637 Å) | | 主引用文献 | Crystal structure of C-terminal domain of putative chitinase from Desulfitobacterium hafniense DCB-2
To Be Published
|
|
4WER
 
 | | Crystal structure of diacylglycerol kinase catalytic domain protein from Enterococcus faecalis V583 | | 分子名称: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, Diacylglycerol kinase catalytic domain protein | | 著者 | Chang, C, Clancy, S, Hatzos-Skintges, C, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2014-09-10 | | 公開日 | 2014-09-24 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Crystal structure of diacylglycerol kinase catalytic domain protein from Enterococcus faecalis V583
To Be Published
|
|
4WHI
 
 | | Crystal structure of C-terminal domain of penicillin binding protein Rv0907 | | 分子名称: | BROMIDE ION, Beta-lactamase, NICKEL (II) ION | | 著者 | Chang, C, Hatzos-Skintges, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2014-09-22 | | 公開日 | 2014-10-08 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure of C-terminal domain of penicillin binding protein Rv0907
To Be Published
|
|
7BYY
 
 | |
6WHF
 
 | | class C beta-lactamase from Escherichia coli in complex with cephalothin | | 分子名称: | (2R)-5-[(acetyloxy)methyl]-2-{(1R)-2-oxo-1-[(thiophen-2-ylacetyl)amino]ethyl}-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase | | 著者 | Chang, C, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-04-08 | | 公開日 | 2020-04-22 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | class C beta-lactamase from Escherichia coli in complex with Cephalothin
To Be Published
|
|
6WIF
 
 | | Class C beta-lactamase from Acinetobacter baumannii in complex with 4-(Ethyl(methyl)carbamoyl)phenyl boronic acid | | 分子名称: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-lactamase, ... | | 著者 | Chang, C, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-04-09 | | 公開日 | 2020-04-22 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | class C beta-lactamase from Acinetobacter baumannii in complex with 4-(Ethyl(methyl)carbamoyl)phenyl boronic acid
To Be Published
|
|
6WJM
 
 | | The crystal structure beta-lactamase from Desulfarculus baarsii DSM 2075 | | 分子名称: | ALANINE, Beta-lactamase, CITRIC ACID | | 著者 | Chang, C, Welk, L, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-04-14 | | 公開日 | 2020-04-22 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | The crystal structure beta-lactamase from Desulfarculus baarsii DSM 2075
To Be Published
|
|
4W66
 
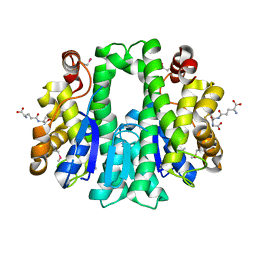 | |
3CWF
 
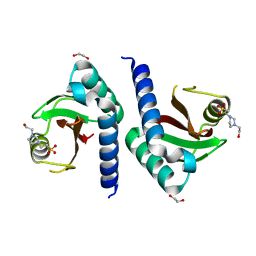 | | Crystal structure of PAS domain of two-component sensor histidine kinase | | 分子名称: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Alkaline phosphatase synthesis sensor protein phoR | | 著者 | Chang, C, Tesar, C, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2008-04-21 | | 公開日 | 2008-05-06 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Extracytoplasmic PAS-like domains are common in signal transduction proteins.
J.Bacteriol., 192, 2010
|
|
5DEQ
 
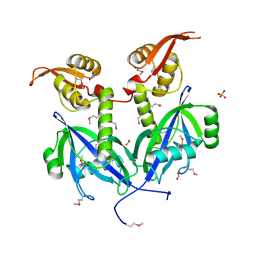 | | Crystal structure of transcriptional factor AraR from Bacteroides thetaiotaomicron VPI in complex with L-arabinose | | 分子名称: | FORMIC ACID, SULFATE ION, TRANSCRIPTIONAL REGULATOR AraR, ... | | 著者 | Chang, C, Tesar, C, Rodionov, D, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2015-08-25 | | 公開日 | 2015-10-21 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | A novel transcriptional regulator of L-arabinose utilization in human gut bacteria.
Nucleic Acids Res., 43, 2015
|
|
1IHR
 
 | | Crystal structure of the dimeric C-terminal domain of TonB | | 分子名称: | BROMIDE ION, TonB protein | | 著者 | Chang, C, Mooser, A, Pluckthun, A, Wlodawer, A. | | 登録日 | 2001-04-20 | | 公開日 | 2001-08-01 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Crystal structure of the dimeric C-terminal domain of TonB reveals a novel fold.
J.Biol.Chem., 276, 2001
|
|
4GAK
 
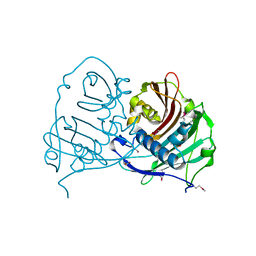 | | Crystal structure of acyl-ACP thioesterase from Spirosoma linguale | | 分子名称: | Acyl-ACP thioesterase, CHLORIDE ION, GLYCEROL | | 著者 | Chang, C, Wu, R, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2012-07-25 | | 公開日 | 2012-09-19 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of acyl-ACP thioesterase from Spirosoma linguale
To be Published
|
|
3D6J
 
 | | Crystal structure of Putative haloacid dehalogenase-like hydrolase from Bacteroides fragilis | | 分子名称: | GLYCEROL, PHOSPHATE ION, Putative haloacid dehalogenase-like hydrolase | | 著者 | Chang, C, Wu, R, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2008-05-19 | | 公開日 | 2008-07-15 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of Putative haloacid dehalogenase-like hydrolase from Bacteroides fragilis
To be Published
|
|
3D1L
 
 | | Crystal structure of putative NADP oxidoreductase BF3122 from Bacteroides fragilis | | 分子名称: | 2-MERCAPTO-PROPION ALDEHYDE, CHLORIDE ION, Putative NADP oxidoreductase BF3122 | | 著者 | Chang, C, Hendricks, R, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2008-05-06 | | 公開日 | 2008-07-08 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | Crystal structure of putative NADP oxidoreductase BF3122 from Bacteroides fragilis.
To be Published
|
|
3CJN
 
 | | Crystal structure of transcriptional regulator, MarR family, from Silicibacter pomeroyi | | 分子名称: | PHOSPHATE ION, Transcriptional regulator, MarR family | | 著者 | Chang, C, Volkart, L, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2008-03-13 | | 公開日 | 2008-03-25 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal structure of MarR family transcriptional regulator from Silicibacter pomeroyi.
To be Published
|
|
5DN1
 
 | | Crystal structure of Phosphoribosyl isomerase A from Streptomyces coelicolor | | 分子名称: | AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, GLYCEROL, Phosphoribosyl isomerase A, ... | | 著者 | Chang, C, Verduzco-Castro, E.A, Endres, M, Barona-Gomez, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2015-09-09 | | 公開日 | 2015-09-30 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.953 Å) | | 主引用文献 | Co-occurrence of analogous enzymes determines evolution of a novel ( beta alpha )8-isomerase sub-family after non-conserved mutations in flexible loop.
Biochem. J., 473, 2016
|
|
3UKJ
 
 | | Crystal structure of extracellular ligand-binding receptor from Rhodopseudomonas palustris HaA2 | | 分子名称: | 3-(4-HYDROXY-PHENYL)PYRUVIC ACID, Extracellular ligand-binding receptor, GLYCEROL, ... | | 著者 | Chang, C, Mack, J, Zerbs, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2011-11-09 | | 公開日 | 2011-11-23 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural and functional characterization of solute binding proteins for aromatic compounds derived from lignin: p-Coumaric acid and related aromatic acids.
Proteins, 81, 2013
|
|
