7KDW
 
 | |
7KDZ
 
 | |
7K6R
 
 | |
2GXB
 
 | | Crystal Structure of The Za Domain bound to Z-RNA | | Descriptor: | 5'-R(P*(DU)P*CP*GP*CP*GP*CP*G)-3', Double-stranded RNA-specific adenosine deaminase, SODIUM ION | | Authors: | Athanasiadis, A, Placido, D, Rich, A. | | Deposit date: | 2006-05-08 | | Release date: | 2007-05-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A Left-Handed RNA Double Helix Bound by the Zalpha Domain of the RNA-Editing Enzyme ADAR1.
Structure, 15, 2007
|
|
1I9S
 
 | | CRYSTAL STRUCTURE OF THE RNA TRIPHOSPHATASE DOMAIN OF MOUSE MRNA CAPPING ENZYME | | Descriptor: | CACODYLATE ION, ISOPROPYL ALCOHOL, MAGNESIUM ION, ... | | Authors: | Changela, A, Ho, C.K, Martins, A, Shuman, S, Mondragon, A. | | Deposit date: | 2001-03-20 | | Release date: | 2001-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and mechanism of the RNA triphosphatase component of mammalian mRNA capping enzyme.
EMBO J., 20, 2001
|
|
1I7D
 
 | | NONCOVALENT COMPLEX OF E.COLI DNA TOPOISOMERASE III WITH AN 8-BASE SINGLE-STRANDED DNA OLIGONUCLEOTIDE | | Descriptor: | 5'-D(*CP*GP*CP*AP*AP*CP*TP*T)-3', CHLORIDE ION, DNA TOPOISOMERASE III, ... | | Authors: | Changela, A, DiGate, R.J, Mondragon, A. | | Deposit date: | 2001-03-08 | | Release date: | 2001-06-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of a complex of a type IA DNA topoisomerase with a single-stranded DNA molecule.
Nature, 411, 2001
|
|
1I9T
 
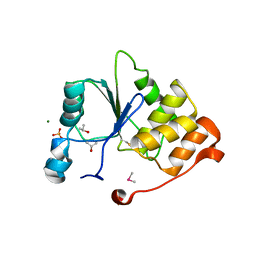 | | CRYSTAL STRUCTURE OF THE OXIDIZED RNA TRIPHOSPHATASE DOMAIN OF MOUSE MRNA CAPPING ENZYME | | Descriptor: | CACODYLATE ION, ISOPROPYL ALCOHOL, MAGNESIUM ION, ... | | Authors: | Changela, A, Ho, C.K, Martins, A, Shuman, S, Mondragon, A. | | Deposit date: | 2001-03-20 | | Release date: | 2001-05-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and mechanism of the RNA triphosphatase component of mammalian mRNA capping enzyme.
EMBO J., 20, 2001
|
|
3RSC
 
 | | Crystal Structure of CalG2, Calicheamicin Glycosyltransferase, TDP and calicheamicin T0 bound form | | Descriptor: | CalG2, Calicheamicin T0, PHOSPHATE ION, ... | | Authors: | Chang, A, Helmich, K.E, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2011-05-02 | | Release date: | 2011-08-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Complete set of glycosyltransferase structures in the calicheamicin biosynthetic pathway reveals the origin of regiospecificity.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
5XMX
 
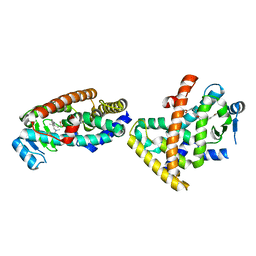 | | Co-crystal structure of Inhibitor compound in complex with human PPARdelta LBD | | Descriptor: | (E)-6-[2-[[[4-(furan-2-yl)phenyl]carbonyl-methyl-amino]methyl]phenoxy]-4-methyl-hex-4-enoic acid, Peroxisome proliferator-activated receptor delta | | Authors: | Lakshminarasimhan, A, Rani, S.T, Senaiar, R.S, Krishnamurthy, N. | | Deposit date: | 2017-05-16 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Novel highly selective peroxisome proliferator-activated receptor delta (PPAR delta) modulators with pharmacokinetic properties suitable for once-daily oral dosing.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
1YN9
 
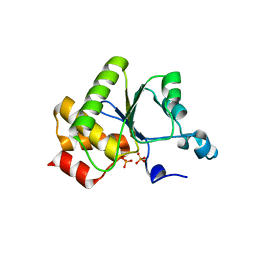 | | Crystal structure of baculovirus RNA 5'-phosphatase complexed with phosphate | | Descriptor: | PHOSPHATE ION, polynucleotide 5'-phosphatase | | Authors: | Changela, A, Martins, A, Shuman, S, Mondragon, A. | | Deposit date: | 2005-01-24 | | Release date: | 2005-02-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of baculovirus RNA triphosphatase complexed with phosphate
J.Biol.Chem., 280, 2005
|
|
6IF7
 
 | | Crystal Structure of AA10 Lytic Polysaccharide Monooxygenase from Tectaria macrodonta | | Descriptor: | COPPER (II) ION, Chitin binding protein, GLYCEROL, ... | | Authors: | Archana, A, Yadav, S.K, Singh, P.K, Vasudev, P.G. | | Deposit date: | 2018-09-18 | | Release date: | 2019-04-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Insecticidal fern protein Tma12 is possibly a lytic polysaccharide monooxygenase.
Planta, 249, 2019
|
|
2H3H
 
 | |
4AP4
 
 | | Rnf4 - ubch5a - ubiquitin heterotrimeric complex | | Descriptor: | E3 UBIQUITIN LIGASE RNF4, UBIQUITIN C, UBIQUITIN-CONJUGATING ENZYME E2 D1, ... | | Authors: | Plechanovova, A, Hay, R.T, Tatham, M.H, Jaffray, E, Naismith, J.H. | | Deposit date: | 2012-03-30 | | Release date: | 2012-07-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure of a Ring E3 Ligase and Ubiquitin-Loaded E2 Primed for Catalysis
Nature, 489, 2012
|
|
2O5C
 
 | | Structure of E. coli topoisomerase III in complex with an 8-base single stranded oligonucleotide. Frozen in glucose pH 5.5 | | Descriptor: | 5'-D(*CP*GP*CP*AP*AP*CP*TP*T)-3', CHLORIDE ION, DNA topoisomerase 3, ... | | Authors: | Changela, A, DiGate, R.J, Mondragon, A. | | Deposit date: | 2006-12-05 | | Release date: | 2007-04-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Studies of E. coli Topoisomerase III-DNA Complexes Reveal a Novel Type IA Topoisomerase-DNA Conformational Intermediate.
J.Mol.Biol., 368, 2007
|
|
2O19
 
 | | Structure of E. coli topoisomersae III in complex with an 8-base single stranded oligonucleotide. Frozen in glycerol at pH 5.5 | | Descriptor: | 5'-D(*CP*GP*CP*AP*AP*CP*TP*T)-3', ACETIC ACID, CHLORIDE ION, ... | | Authors: | Changela, A, DiGate, R, Mondragon, A. | | Deposit date: | 2006-11-28 | | Release date: | 2007-04-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural Studies of E. coli Topoisomerase III-DNA Complexes Reveal a Novel Type IA Topoisomerase-DNA Conformational Intermediate.
J.Mol.Biol., 368, 2007
|
|
2O54
 
 | | Structure of E. coli topoisomerase III in complex with an 8-base single stranded oligonucleotide. Frozen in glycerol at pH 7.0 | | Descriptor: | 5'-D(*CP*GP*CP*AP*AP*CP*TP*T)-3', ACETIC ACID, CHLORIDE ION, ... | | Authors: | Changela, A, Digate, R.J, Mondragon, A. | | Deposit date: | 2006-12-05 | | Release date: | 2007-04-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Studies of E. coli Topoisomerase III-DNA Complexes Reveal a Novel Type IA Topoisomerase-DNA Conformational Intermediate.
J.Mol.Biol., 368, 2007
|
|
2O5E
 
 | | Structure of E. coli topoisomerase III in complex with an 8-base single stranded oligonucleotide. Frozen in glucose pH 7.0 | | Descriptor: | 5'-D(*CP*GP*CP*AP*AP*CP*TP*T)-3', CHLORIDE ION, DNA topoisomerase 3, ... | | Authors: | Changela, A, DiGate, R.J, Mondragon, A. | | Deposit date: | 2006-12-05 | | Release date: | 2007-04-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Studies of E. coli Topoisomerase III-DNA Complexes Reveal a Novel Type IA Topoisomerase-DNA Conformational Intermediate.
J.Mol.Biol., 368, 2007
|
|
9CZ1
 
 | |
1XMK
 
 | | The Crystal structure of the Zb domain from the RNA editing enzyme ADAR1 | | Descriptor: | CADMIUM ION, CHLORIDE ION, Double-stranded RNA-specific adenosine deaminase, ... | | Authors: | Athanasiadis, A, Placido, D, Maas, S, Brown II, B.A, Lowenhaupt, K, Rich, A. | | Deposit date: | 2004-10-03 | | Release date: | 2005-08-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | The Crystal Structure of the Z[beta] Domain of the RNA-editing Enzyme ADAR1 Reveals Distinct Conserved Surfaces Among Z-domains.
J.Mol.Biol., 351, 2005
|
|
9CZ2
 
 | |
3TSR
 
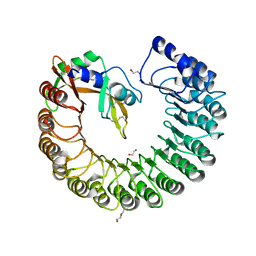 | | X-ray structure of mouse ribonuclease inhibitor complexed with mouse ribonuclease 1 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Ribonuclease inhibitor, ... | | Authors: | Chang, A, Lomax, J.E, Bingman, C.A, Raines, R.T, Phillips Jr, G.N. | | Deposit date: | 2011-09-13 | | Release date: | 2012-09-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1999 Å) | | Cite: | Functional evolution of ribonuclease inhibitor: insights from birds and reptiles.
J.Mol.Biol., 426, 2014
|
|
3TOS
 
 | | Crystal Structure of CalS11, Calicheamicin Methyltransferase | | Descriptor: | 1,2-ETHANEDIOL, CalS11, GLUTAMIC ACID, ... | | Authors: | Chang, A, Aceti, D.J, Beebe, E.T, Makino, S.-I, Wrobel, R.L, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2011-09-06 | | Release date: | 2011-10-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of CalS11, Calicheamicin methyltransferase
To be Published
|
|
2O59
 
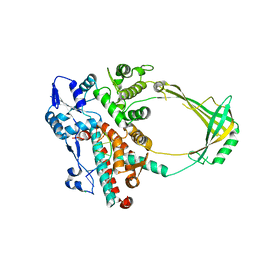 | | Structure of E. coli topoisomerase III in complex with an 8-base single stranded oligonucleotide. Frozen in glycerol pH 8.0 | | Descriptor: | 5'-D(*CP*GP*CP*AP*AP*CP*TP*T)-3', ACETIC ACID, CHLORIDE ION, ... | | Authors: | Changela, A, DiGate, R.J, Mondragon, A. | | Deposit date: | 2006-12-05 | | Release date: | 2007-04-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Studies of E. coli Topoisomerase III-DNA Complexes Reveal a Novel Type IA Topoisomerase-DNA Conformational Intermediate.
J.Mol.Biol., 368, 2007
|
|
4JMQ
 
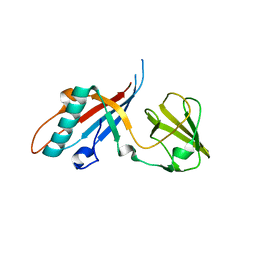 | | Crystal structure of pb9: The Dit of bacteriophage T5. | | Descriptor: | Bacteriophage T5 distal tail protein | | Authors: | Flayhan, A, Vellieux, F.M.D, Girard, E, Maury, O, Boulanger, P, Breyton, C. | | Deposit date: | 2013-03-14 | | Release date: | 2013-11-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Crystal Structure of pb9, the Distal Tail Protein of Bacteriophage T5: a Conserved Structural Motif among All Siphophages.
J.Virol., 88, 2014
|
|
1IAZ
 
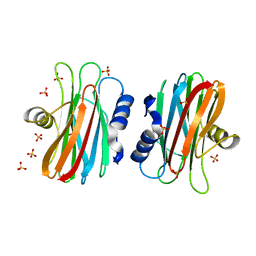 | | EQUINATOXIN II | | Descriptor: | EQUINATOXIN II, SULFATE ION | | Authors: | Athanasiadis, A, Anderluh, G, Macek, P, Turk, D. | | Deposit date: | 2001-03-24 | | Release date: | 2001-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the soluble form of equinatoxin II, a pore-forming toxin from the sea anemone Actinia equina.
Structure, 9, 2001
|
|
