4E5T
 
 | | Crystal structure of a putative Mandelate racemase/Muconate lactonizing enzyme (Target PSI-200750) from Labrenzia alexandrii DFL-11 | | Descriptor: | MAGNESIUM ION, Mandelate racemase / muconate lactonizing enzyme, C-terminal domain protein | | Authors: | Kumar, P.R, Bonanno, J, Chowdhury, S, Foti, R, Gizzi, A, Glen, S, Hammonds, J, Hillerich, B, Matikainen, B, Seidel, R, Toro, R, Zencheck, W, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-03-14 | | Release date: | 2012-03-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of a putative MR/ML enzyme from Labrenzia alexandrii DFL-11
to be published
|
|
1KZX
 
 | | Solution structure of human intestinal fatty acid binding protein with a naturally-occurring single amino acid substitution (A54T) | | Descriptor: | INTESTINAL FATTY ACID-BINDING PROTEIN (T54) | | Authors: | Zhang, F, Luecke, C, Baier, L.J, Sacchettini, J.C, Hamilton, J.A. | | Deposit date: | 2002-02-08 | | Release date: | 2003-07-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human intestinal fatty acid binding protein with a naturally-occurring single amino acid substitution (A54T) that is associated with altered lipid metabolism
Biochemistry, 42, 2003
|
|
3BBX
 
 | |
2J6V
 
 | | Crystal structure of the DNA repair enzyme UV Damage Endonuclease | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, UV ENDONUCLEASE | | Authors: | Paspaleva, K, Thomassen, E.A.J, Pannu, N.S, Goossen, N, Abrahams, J.P. | | Deposit date: | 2006-10-04 | | Release date: | 2007-10-16 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of the DNA Repair Enzyme Ultraviolet Damage Endonuclease.
Structure, 15, 2007
|
|
4NNP
 
 | |
4MDC
 
 | | Crystal structure of glutathione S-transferase from Sinorhizobium meliloti 1021, NYSGRC target 021389 | | Descriptor: | GLYCEROL, Putative glutathione S-transferase | | Authors: | Shabalin, I.G, Bacal, P, Cooper, D.R, Stead, M, Ahmed, M, Hammonds, J, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-08-22 | | Release date: | 2013-09-04 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of glutathione S-transferase from Sinorhizobium meliloti 1021
To be Published
|
|
1RFI
 
 | | Crystal structure of human Tyrosyl-DNA Phosphodiesterase complexed with vanadate, pentapeptide KLNYK, and tetranucleotide AGTC | | Descriptor: | 5'-D(*AP*GP*TP*C)-3', SPERMINE, Topoisomerase I-Derived Peptide, ... | | Authors: | Davies, D.R, Interthal, H, Champoux, J.J, Hol, W.G. | | Deposit date: | 2003-11-10 | | Release date: | 2004-03-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Explorations of peptide and oligonucleotide binding sites of tyrosyl-DNA phosphodiesterase using vanadate complexes.
J.Med.Chem., 47, 2004
|
|
3BBV
 
 | |
1RFF
 
 | | Crystal structure of human Tyrosyl-DNA Phosphodiesterase complexed with vanadate, octapeptide KLNYYDPR, and tetranucleotide AGTT. | | Descriptor: | 5'-D(*AP*GP*TP*T)-3', SPERMINE, Topoisomerase I-Derived Peptide, ... | | Authors: | Davies, D.R, Interthal, H, Champoux, J.J, Hol, W.G. | | Deposit date: | 2003-11-08 | | Release date: | 2004-03-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Explorations of peptide and oligonucleotide binding sites of tyrosyl-DNA phosphodiesterase using vanadate complexes.
J.Med.Chem., 47, 2004
|
|
1RG2
 
 | | Crystal structure of human Tyrosyl-DNA Phosphodiesterase complexed with vanadate, octopamine, and tetranucleotide AGTA | | Descriptor: | 4-(2R-AMINO-1-HYDROXYETHYL)PHENOL, 4-(2S-AMINO-1-HYDROXYETHYL)PHENOL, 5'-D(*AP*GP*TP*A)-3', ... | | Authors: | Davies, D.R, Interthal, H, Champoux, J.J, Hol, W.G. | | Deposit date: | 2003-11-11 | | Release date: | 2004-03-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Explorations of peptide and oligonucleotide binding sites of tyrosyl-DNA phosphodiesterase using vanadate complexes.
J.Med.Chem., 47, 2004
|
|
2LKK
 
 | | Human L-FABP in complex with oleate | | Descriptor: | Fatty acid-binding protein, liver, OLEIC ACID | | Authors: | Cai, J, Luecke, C, Chen, Z, Qiao, Y, Klimtchuk, E.S, Hamilton, J.A. | | Deposit date: | 2011-10-13 | | Release date: | 2012-06-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of human liver fatty acid binding protein: fatty acid binding revisited.
Biophys.J., 102, 2012
|
|
1MII
 
 | | SOLUTION STRUCTURE OF ALPHA-CONOTOXIN MII | | Descriptor: | PROTEIN (ALPHA CONOTOXIN MII) | | Authors: | Hill, J.M, Oomen, C.J, Miranda, L.P, Bingham, J.P, Alewood, P.F, Craik, D.J. | | Deposit date: | 1998-10-05 | | Release date: | 1998-10-21 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of alpha-conotoxin MII by NMR spectroscopy: effects of solution environment on helicity.
Biochemistry, 37, 1998
|
|
4Q60
 
 | | Crystal structure of a 4-hydroxyproline epimerase from Burkholderia Multivorans atcc 17616, target EFI-506586, open form, with bound pyrrole-2-carboxylate | | Descriptor: | GLYCEROL, PROLINE RACEMASE, PYRROLE-2-CARBOXYLATE | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Sojitra, S, Stead, M, Washington, E, Glenn, A.S, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-04-18 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | CRYSTAL STRUCTURE OF PROLINE RACEMASE Bmul_4447 FROM Burkholderia multivorans, TARGET EFI-506586
To be Published
|
|
1RG1
 
 | | Crystal structure of human Tyrosyl-DNA Phosphodiesterase complexed with vanadate, octopamine, and tetranucleotide AGTT | | Descriptor: | 4-(2S-AMINO-1-HYDROXYETHYL)PHENOL, 5'-D(*AP*GP*TP*T)-3', SPERMINE, ... | | Authors: | Davies, D.R, Interthal, H, Champoux, J.J, Hol, W.G. | | Deposit date: | 2003-11-10 | | Release date: | 2004-03-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Explorations of peptide and oligonucleotide binding sites of tyrosyl-DNA phosphodiesterase using vanadate complexes.
J.Med.Chem., 47, 2004
|
|
1RGT
 
 | | Crystal structure of human Tyrosyl-DNA Phosphodiesterase complexed with vanadate, octopamine, and tetranucleotide AGTC | | Descriptor: | 4-(2R-AMINO-1-HYDROXYETHYL)PHENOL, 4-(2S-AMINO-1-HYDROXYETHYL)PHENOL, 5'-D(*AP*GP*TP*C)-3', ... | | Authors: | Davies, D.R, Interthal, H, Champoux, J.J, Hol, W.G. | | Deposit date: | 2003-11-12 | | Release date: | 2004-03-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Explorations of peptide and oligonucleotide binding sites of tyrosyl-DNA phosphodiesterase using vanadate complexes.
J.Med.Chem., 47, 2004
|
|
1RGU
 
 | | The crystal structure of human Tyrosyl-DNA Phosphodiesterase complexed with vanadate, octopamine, and tetranucleotide AGTG | | Descriptor: | 4-(2S-AMINO-1-HYDROXYETHYL)PHENOL, 5'-D(*AP*GP*TP*G)-3', SPERMINE, ... | | Authors: | Davies, D.R, Interthal, H, Champoux, J.J, Hol, W.G. | | Deposit date: | 2003-11-13 | | Release date: | 2004-03-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Explorations of peptide and oligonucleotide binding sites of tyrosyl-DNA phosphodiesterase using vanadate complexes.
J.Med.Chem., 47, 2004
|
|
4OKH
 
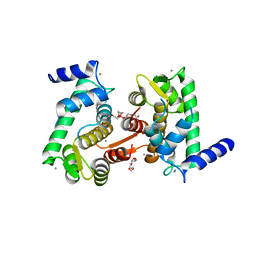 | | Crystal structure of calpain-3 penta-EF-hand domain | | Descriptor: | 2,5,8,11,14,17,20,23,26,29,32,35,38,41,44,47,50,53,56,59,62,65,68,71,74,77,80-HEPTACOSAOXADOOCTACONTAN-82-OL, CALCIUM ION, Calpain-3 | | Authors: | Karunan Partha, S, Ravulapalli, R, Campbell, R.L, Allingham, J.S, Davies, P.L. | | Deposit date: | 2014-01-22 | | Release date: | 2014-05-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of calpain-3 penta-EF-hand (PEF) domain - a homodimerized PEF family member with calcium bound at the fifth EF-hand.
Febs J., 281, 2014
|
|
2L68
 
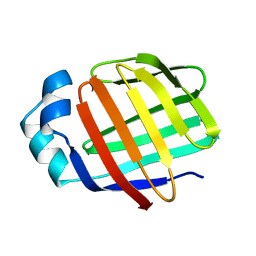 | | Solution Structure of Human Holo L-FABP | | Descriptor: | Fatty acid-binding protein, liver | | Authors: | Cai, J, Luecke, C, Chen, Z, Qiao, Y, Klimtchuk, E.S, Hamilton, J.A. | | Deposit date: | 2010-11-17 | | Release date: | 2011-11-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of human liver fatty acid binding protein: fatty acid binding revisited.
Biophys.J., 102, 2012
|
|
1RH0
 
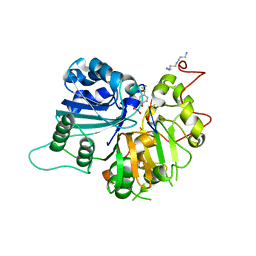 | | Crystal structure of human Tyrosyl-DNA Phosphodiesterase complexed with vanadate, octopamine and trinucleotide GTT | | Descriptor: | 4-(2S-AMINO-1-HYDROXYETHYL)PHENOL, 5'-D(*GP*TP*T)-3', SPERMINE, ... | | Authors: | Davies, D.R, Interthal, H, Champoux, J.J, Hol, W.G. | | Deposit date: | 2003-11-13 | | Release date: | 2004-03-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Explorations of peptide and oligonucleotide binding sites of tyrosyl-DNA phosphodiesterase using vanadate complexes.
J.Med.Chem., 47, 2004
|
|
1ZNI
 
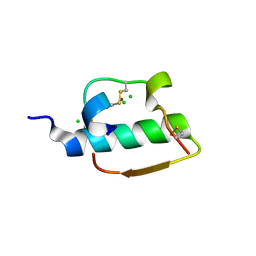 | | INSULIN | | Descriptor: | CHLORIDE ION, INSULIN, ZINC ION | | Authors: | Turkenburg, M.G.W, Whittingham, J.L, Dodson, G.G, Dodson, E.J, Xiao, B, Bentley, G.A. | | Deposit date: | 1997-09-23 | | Release date: | 1998-01-28 | | Last modified: | 2018-07-04 | | Method: | X-RAY DIFFRACTION (1.498 Å) | | Cite: | Structure of insulin in 4-zinc insulin.
Nature, 261, 1976
|
|
1BR8
 
 | | IMPLICATIONS FOR FUNCTION AND THERAPY OF A 2.9A STRUCTURE OF BINARY-COMPLEXED ANTITHROMBIN | | Descriptor: | PROTEIN (ANTITHROMBIN-III), PROTEIN (PEPTIDE) | | Authors: | Skinner, R, Chang, W.S.W, Jin, L, Pei, X.Y, Huntington, J.A, Abrahams, J.P, Carrell, R.W, Lomas, D.A. | | Deposit date: | 1998-08-26 | | Release date: | 1998-09-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Implications for function and therapy of a 2.9 A structure of binary-complexed antithrombin.
J.Mol.Biol., 283, 1998
|
|
2L67
 
 | | Solution Structure of Human Apo L-FABP | | Descriptor: | Fatty acid-binding protein, liver | | Authors: | Cai, J, Luecke, C, Chen, Z, Qiao, Y, Klimtchuk, E.S, Hamilton, J.A. | | Deposit date: | 2010-11-17 | | Release date: | 2011-11-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of human liver fatty acid binding protein: fatty acid binding revisited.
Biophys.J., 102, 2012
|
|
3VA8
 
 | | Crystal structure of enolase FG03645.1 (target EFI-502278) from Gibberella zeae PH-1 complexed with magnesium, formate and sulfate | | Descriptor: | FORMIC ACID, MAGNESIUM ION, PROBABLE DEHYDRATASE, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-12-29 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of dehydratase FG03645.1 from Gibberella zeae PH-1
To be Published
|
|
3VC5
 
 | | Crystal structure of enolase Tbis_1083(TARGET EFI-502310) FROM Thermobispora bispora DSM 43833 complexed with phosphate | | Descriptor: | Mandelate racemase/muconate lactonizing protein, PHOSPHATE ION | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-01-03 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of enolase Tbis_1083 FROM Thermobispora bispora DSM 43833
To be Published
|
|
3VDG
 
 | | Crystal structure of enolase MSMEG_6132 (TARGET EFI-502282) from Mycobacterium smegmatis str. MC2 155 complexed with formate and acetate | | Descriptor: | ACETATE ION, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-01-05 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of enolase MSMEG_6132 FROM Mycobacterium smegmatis
To be Published
|
|
