1YVH
 
 | |
2AXL
 
 | | Solution structure of a multifunctional DNA- and protein-binding domain of human Werner syndrome protein | | Descriptor: | Werner syndrome | | Authors: | Hu, J.-S, Feng, H, Zeng, W, Lin, G.-X, Xi, X.G. | | Deposit date: | 2005-09-05 | | Release date: | 2005-12-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a multifunctional DNA- and protein-binding motif of human Werner syndrome protein.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
8I99
 
 | | N-carbamoyl-D-amino-acid hydrolase mutant - M4Th3 | | Descriptor: | N-carbamoyl-D-amino-acid hydrolase | | Authors: | Hu, J.M, Ni, Y, Xu, G.C. | | Deposit date: | 2023-02-06 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Engineering the Thermostability of a d-Carbamoylase Based on Ancestral Sequence Reconstruction for the Efficient Synthesis of d-Tryptophan.
J.Agric.Food Chem., 71, 2023
|
|
2HDX
 
 | |
5YAK
 
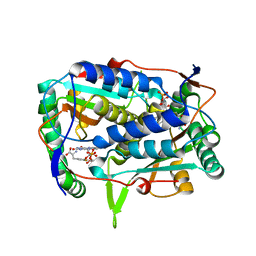 | |
2H95
 
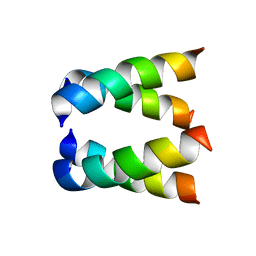 | |
2HDV
 
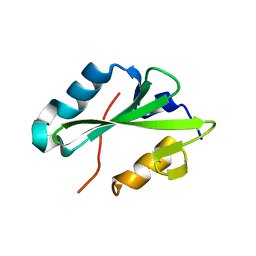 | |
6DWB
 
 | | Structure of the Salmonella SPI-1 type III secretion injectisome needle filament | | Descriptor: | Protein PrgI | | Authors: | Hu, J, Hong, C, Worrall, L.J, Vuckovic, M, Yu, Z, Strynadka, N.C.J. | | Deposit date: | 2018-06-26 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM analysis of the T3S injectisome reveals the structure of the needle and open secretin.
Nat Commun, 9, 2018
|
|
6DV3
 
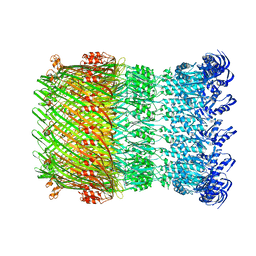 | | Structure of the Salmonella SPI-1 type III secretion injectisome secretin InvG in the open gate state | | Descriptor: | Protein InvG | | Authors: | Hu, J, Worrall, L.J, Vuckovic, M, Atkinson, C.E, Strynadka, N.C.J. | | Deposit date: | 2018-06-22 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM analysis of the T3S injectisome reveals the structure of the needle and open secretin.
Nat Commun, 9, 2018
|
|
7X4B
 
 | | Crystal Structure of An Anti-CRISPR Protein | | Descriptor: | Anti-CRISPR protein (AcrIIC1), SULFATE ION | | Authors: | Hu, J, Zhang, S, Gao, J.Y, Liu, X, Liu, J. | | Deposit date: | 2022-03-02 | | Release date: | 2022-10-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | A redox switch regulates the assembly and anti-CRISPR activity of AcrIIC1.
Nat Commun, 13, 2022
|
|
5TDE
 
 | | TEV Cleaved Human ATP Citrate Lyase Bound to Citrate | | Descriptor: | ATP-citrate synthase, CITRATE ANION, DIHYDROGENPHOSPHATE ION, ... | | Authors: | Hu, J, Fraser, M.E. | | Deposit date: | 2016-09-19 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Binding of hydroxycitrate to human ATP-citrate lyase.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5TDZ
 
 | |
5TEQ
 
 | |
5TDF
 
 | |
5TE1
 
 | |
5TDM
 
 | |
5TES
 
 | |
5TET
 
 | |
3S0X
 
 | |
5YDU
 
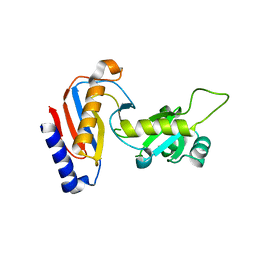 | | Crystal structure of Utp30 | | Descriptor: | PHOSPHATE ION, Ribosome biogenesis protein UTP30 | | Authors: | Hu, J, Zhu, X, Ye, K. | | Deposit date: | 2017-09-14 | | Release date: | 2017-11-01 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.646 Å) | | Cite: | Structure and RNA recognition of ribosome assembly factor Utp30.
RNA, 23, 2017
|
|
7CQZ
 
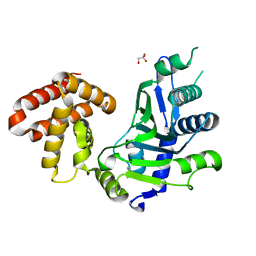 | | crystal structure of mouse FAM46C (TENT5C) | | Descriptor: | CHLORIDE ION, GLYCEROL, Terminal nucleotidyltransferase 5C | | Authors: | Hu, J.L, Zhang, H, Gao, S. | | Deposit date: | 2020-08-12 | | Release date: | 2021-06-09 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.34969759 Å) | | Cite: | Structural and functional characterization of multiple myeloma associated cytoplasmic poly(A) polymerase FAM46C.
Cancer Commun (Lond), 41, 2021
|
|
7MJ1
 
 | | LarB, a carboxylase/hydrolase involved in synthesis of the cofactor for lactate racemase, in complex with NAD | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Chatterjee, S, Rankin, J.A, Lagishetty, S, Hu, J, Hausinger, R.P. | | Deposit date: | 2021-04-19 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.402 Å) | | Cite: | The LarB carboxylase/hydrolase forms a transient cysteinyl-pyridine intermediate during nickel-pincer nucleotide cofactor biosynthesis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MJ2
 
 | | LarB, a carboxylase/hydrolase involved in synthesis of the cofactor for lactate racemase, in complex with Zn | | Descriptor: | MAGNESIUM ION, Pyridinium-3,5-biscarboxylic acid mononucleotide synthase, ZINC ION | | Authors: | Chatterjee, S, Rankin, J.A, Lagishetty, S, Hu, J, Hausinger, R.P. | | Deposit date: | 2021-04-19 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The LarB carboxylase/hydrolase forms a transient cysteinyl-pyridine intermediate during nickel-pincer nucleotide cofactor biosynthesis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MJ0
 
 | | LarB, a carboxylase/hydrolase involved in synthesis of the cofactor for lactate racemase, in complex with adenosine monophosphate AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, Pyridinium-3,5-biscarboxylic acid mononucleotide synthase | | Authors: | Chatterjee, S, Rankin, J.A, Lagishetty, S, Hu, J, Hausinger, R.P. | | Deposit date: | 2021-04-19 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | The LarB carboxylase/hydrolase forms a transient cysteinyl-pyridine intermediate during nickel-pincer nucleotide cofactor biosynthesis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1VSQ
 
 | |
