7LSR
 
 | | Ruminococcus bromii Amy12-D392A with maltoheptaose | | Descriptor: | CALCIUM ION, GLYCEROL, Pullulanase, ... | | Authors: | Koropatkin, N.M, Cockburn, D.W, Brown, H.A, Kibler, R.D. | | Deposit date: | 2021-02-18 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structure and substrate recognition by the Ruminococcus bromii amylosome pullulanases.
J.Struct.Biol., 213, 2021
|
|
6RV9
 
 | | Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor inactive S270A variant in complex with the aldouronic acid XUXXR | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-O-methyl-alpha-D-glucopyranuronic acid-(1-2)-[beta-D-xylopyranose-(1-4)]beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-Xylitol, ... | | Authors: | Ernst, H.A, Mosbech, C, Langkilde, A, Westh, P, Meyer, A, Agger, J.W, Larsen, S. | | Deposit date: | 2019-05-31 | | Release date: | 2020-03-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | The structural basis of fungal glucuronoyl esterase activity on natural substrates.
Nat Commun, 11, 2020
|
|
6RU2
 
 | | Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-O-methyl-glucuronoyl methylesterase, ... | | Authors: | Ernst, H.A, Mosbech, C, Langkilde, A, Westh, P, Meyer, A, Agger, J.W, Larsen, S. | | Deposit date: | 2019-05-27 | | Release date: | 2020-03-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The structural basis of fungal glucuronoyl esterase activity on natural substrates.
Nat Commun, 11, 2020
|
|
5UG3
 
 | | NMR SOLUTION STRUCTURE OF ALPHA-CONOTOXIN GID MUTANT A10V | | Descriptor: | Alpha-conotoxin GID | | Authors: | Hussein, A.K, Leffler, A.E, Zebroski, H.A, Powell, S.R, Kuryatov, A, Filipenko, P, Gorson, J, Heizmann, A, Lyskov, S, Nicke, A, Lindstrom, J, Rudy, B, Bonneau, R, Holford, M, Poget, S.F. | | Deposit date: | 2017-01-06 | | Release date: | 2017-09-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Discovery of peptide ligands through docking and virtual screening at nicotinic acetylcholine receptor homology models.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5UG5
 
 | | NMR SOLUTION STRUCTURE OF THE ALPHA-CONOTOXIN GID MUTANT V13Y | | Descriptor: | Alpha-conotoxin GID | | Authors: | Hussein, A, Leffler, A.E, Kuryatov, A, Zebroski, H.A, Powell, S.R, Filipenko, P, Gorson, J, Heizmann, A, Lyskov, S, Nicke, A, Lindstrom, J, Rudy, B, Bonneau, R, Holford, M, Poget, S.F. | | Deposit date: | 2017-01-06 | | Release date: | 2017-09-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Discovery of peptide ligands through docking and virtual screening at nicotinic acetylcholine receptor homology models.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7NTH
 
 | | Structure of TAK1 in complex with compound 54 | | Descriptor: | 2-[[5-[[2-[bis(fluoranyl)methoxy]phenyl]methyl-[(2~{R})-1-(methylamino)-1-oxidanylidene-propan-2-yl]carbamoyl]-1~{H}-imidazol-2-yl]carbonyl]isoindole-5-carboxamide, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Veerman, J.J.N, Bruseker, Y.B, Damen, E, Heijne, E.H, van Bruggen, W, Hekking, K.F.W, Winkel, R, Hupp, C.D, Keefe, A.D, Liu, J, Thomson, H.A, Zhang, Y, Cuozzo, J.W, McRiner, A.J, Mulvihill, M.J, van Rijnsbergen, P, Zech, B, Renzetti, L.M, Babiss, L, Mueller, G. | | Deposit date: | 2021-03-09 | | Release date: | 2021-04-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Discovery of 2,4-1 H -Imidazole Carboxamides as Potent and Selective TAK1 Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
7NTI
 
 | | Structure of TAK1 in complex with compound 22 | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, Mitogen-activated protein kinase 7,TGF-beta-activated kinase 1 and MAP3K7-binding protein 1, ... | | Authors: | Veerman, J.J.N, Bruseker, Y.B, Damen, E, Heijne, E.H, van Bruggen, W, Hekking, K.F.W, Winkel, R, Hupp, C.D, Keefe, A.D, Liu, J, Thomson, H.A, Zhang, Y, Cuozzo, J.W, McRiner, A.J, Mulvihill, M.J, van Rijnsbergen, P, Zech, B, Renzetti, L.M, Babiss, L, Mueller, G. | | Deposit date: | 2021-03-09 | | Release date: | 2021-04-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Discovery of 2,4-1 H -Imidazole Carboxamides as Potent and Selective TAK1 Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
6EL8
 
 | | Crystal structure of the Forkhead domain of human FOXN1 in complex with DNA | | Descriptor: | DNA (5'-D(*GP*GP*TP*GP*GP*CP*GP*TP*CP*TP*TP*CP*A)-3'), DNA (5'-D(*TP*GP*AP*AP*GP*AP*CP*GP*CP*CP*AP*CP*C)-3'), Forkhead box protein N1 | | Authors: | Newman, J.A, Aitkenhead, H.A, Pinkas, D.M, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-09-28 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Crystal structure of the Forkhead domain of human FOXN1 in complex with DNA
To be published
|
|
5TID
 
 | | X-ray structure of acyl-CoA thioesterase I, TesA, mutant M141L/Y145K/L146K at pH 5 in complex with octanoic acid | | Descriptor: | Acyl-CoA thioesterase I, OCTANOIC ACID (CAPRYLIC ACID) | | Authors: | Thoden, J.B, Holden, H.M, Grisewood, M.J, Hernandez Lozada, N.J, Gifford, N.P, Mendez-Perez, D, Schoenberger, H.A, Allan, M.F, Pfleger, B.F, Marines, C.D. | | Deposit date: | 2016-10-02 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Computational Redesign of Acyl-ACP Thioesterase with Improved Selectivity toward Medium-Chain-Length Fatty Acids.
ACS Catal, 7, 2017
|
|
6GB1
 
 | | Crystal structure of the GLP1 receptor ECD with Peptide 11 | | Descriptor: | Glucagon-like peptide 1 receptor, HEXANE-1,6-DIOL, Peptide 11, ... | | Authors: | Schreuder, H.A, Liesum, A. | | Deposit date: | 2018-04-13 | | Release date: | 2018-06-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Dual Glucagon-like Peptide 1 (GLP-1)/Glucagon Receptor Agonists Specifically Optimized for Multidose Formulations.
J. Med. Chem., 61, 2018
|
|
5TIC
 
 | | X-ray structure of wild-type E. coli Acyl-CoA thioesterase I at pH 5 | | Descriptor: | Acyl-CoA thioesterase I, CHLORIDE ION | | Authors: | Thoden, J.B, Holden, H.M, Grisewood, M.J, Hernandez Lozada, N.J, Gifford, N.P, Mendez-Perez, D, Schoenberger, H.A, Allan, M.F, Pfleger, B.F, Marines, C.D. | | Deposit date: | 2016-10-02 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Computational Redesign of Acyl-ACP Thioesterase with Improved Selectivity toward Medium-Chain-Length Fatty Acids.
ACS Catal, 7, 2017
|
|
6AZU
 
 | | Holo IDO1 crystal structure | | Descriptor: | Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Lewis, H.A, Yan, C. | | Deposit date: | 2017-09-13 | | Release date: | 2018-03-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.822 Å) | | Cite: | Immune-modulating enzyme indoleamine 2,3-dioxygenase is effectively inhibited by targeting its apo-form.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6AZV
 
 | | IDO1/BMS-978587 crystal structure | | Descriptor: | (1R,2S)-2-(4-[bis(2-methylpropyl)amino]-3-{[(4-methylphenyl)carbamoyl]amino}phenyl)cyclopropane-1-carboxylic acid, Indoleamine 2,3-dioxygenase 1 | | Authors: | Lewis, H.A. | | Deposit date: | 2017-09-13 | | Release date: | 2018-03-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.755 Å) | | Cite: | Immune-modulating enzyme indoleamine 2,3-dioxygenase is effectively inhibited by targeting its apo-form.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5V7C
 
 | |
6AQS
 
 | | Crystal structure of Plasmodium falciparum purine nucleoside phosphorylase (V181D) mutant complexed with DADMe-ImmG and phosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-7-{[(3R,4R)-3-hydroxy-4-(hydroxymethyl)pyrrolidin-1-yl]methyl}-3,5-dihydro-4H-pyrrolo[3,2-d]pyrimidin-4-one, PHOSPHATE ION, ... | | Authors: | Harijan, R.K, Ducati, R.G, Namanja-Magliano, H.A, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2017-08-21 | | Release date: | 2018-02-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Genetic resistance to purine nucleoside phosphorylase inhibition in
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2CSA
 
 | | Structure of the M3 Muscarinic Acetylcholine Receptor Basolateral Sorting Signal | | Descriptor: | Muscarinic acetylcholine receptor M3 | | Authors: | Iverson, H.A, Fox, D, Nadler, L.S, Klevit, R.E, Nathanson, N.M. | | Deposit date: | 2005-05-21 | | Release date: | 2005-05-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Identification and structural determination of the M3 muscarinic acetylcholine receptor basolateral sorting signal.
J.Biol.Chem., 280, 2005
|
|
4PTC
 
 | | Structure of a carboxamide compound (3) (2-{2-[(CYCLOPROPYLCARBONYL)AMINO]PYRIDIN-4-YL}-4-OXO-4H-1LAMBDA~4~,3-THIAZOLE-5-CARBOXAMIDE) to GSK3b | | Descriptor: | 2-[2-(cyclopropylcarbonylamino)pyridin-4-yl]-4-methoxy-1,3-thiazole-5-carboxamide, Glycogen synthase kinase-3 beta | | Authors: | Lewis, H.A, Sivaprakasam, P, Kish, K, Pokross, M, Dubowchik, G.M. | | Deposit date: | 2014-03-10 | | Release date: | 2015-04-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.711 Å) | | Cite: | Discovery of new acylaminopyridines as GSK-3 inhibitors by a structure guided in-depth exploration of chemical space around a pyrrolopyridinone core.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
2D3O
 
 | | Structure of Ribosome Binding Domain of the Trigger Factor on the 50S ribosomal subunit from D. radiodurans | | Descriptor: | 23S RIBOSOMAL RNA, 50S RIBOSOMAL PROTEIN L23, 50S RIBOSOMAL PROTEIN L24, ... | | Authors: | Schluenzen, F, Wilson, D.N, Hansen, H.A, Tian, P, Harms, J.M, McInnes, S.J, Albrecht, R, Buerger, J, Wilbanks, S.M, Fucini, P. | | Deposit date: | 2005-09-30 | | Release date: | 2005-12-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | The Binding Mode of the Trigger Factor on the Ribosome: Implications for Protein Folding and SRP Interaction
Structure, 13, 2005
|
|
7TQ7
 
 | | Structure of MERS 3CL protease in complex with the cyclopropane based inhibitor 13c | | Descriptor: | N-{(2S)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-N~2~-({[(1R,2R)-2-propylcyclopropyl]methoxy}carbonyl)-L-leucinamide, Orf1a protein, TETRAETHYLENE GLYCOL | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-01-26 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
7TQ6
 
 | | Structure of SARS-CoV-2 3CL protease in complex with the cyclopropane based inhibitor 13d | | Descriptor: | (1R,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-{[N-({[(1R,2R)-2-propylcyclopropyl]methoxy}carbonyl)-L-leucyl]amino}propane-1-sulfonic acid, (1S,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-{[N-({[(1R,2R)-2-propylcyclopropyl]methoxy}carbonyl)-L-leucyl]amino}propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-01-26 | | Release date: | 2022-02-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
4PTE
 
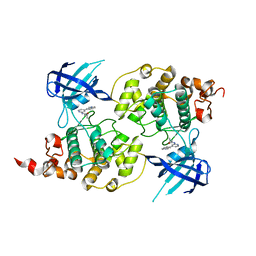 | | Structure of a carvoxamide compound (15) (N-[4-(ISOQUINOLIN-7-YL)PYRIDIN-2-YL]CYCLOPROPANECARBOXAMIDE) to GSK3b | | Descriptor: | Glycogen synthase kinase-3 beta, N-[4-(isoquinolin-7-yl)pyridin-2-yl]cyclopropanecarboxamide | | Authors: | Lewis, H.A, Sivaprakasam, P, Kish, K, Pokross, M, Dubowchik, G.M. | | Deposit date: | 2014-03-10 | | Release date: | 2015-04-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.033 Å) | | Cite: | Discovery of new acylaminopyridines as GSK-3 inhibitors by a structure guided in-depth exploration of chemical space around a pyrrolopyridinone core.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
7TQ5
 
 | | Structure of SARS-CoV-2 3CL protease in complex with the cyclopropane based inhibitor 10d | | Descriptor: | (1R,2S)-1-hydroxy-2-{[N-({[(1R,2R)-2-(4-methoxyphenyl)cyclopropyl]methoxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-1-hydroxy-2-{[N-({[(1R,2R)-2-(4-methoxyphenyl)cyclopropyl]methoxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-01-26 | | Release date: | 2022-02-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
4PTG
 
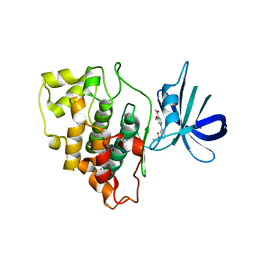 | | Structure of a carboxamine compound (26) (2-{2-[(CYCLOPROPYLCARBONYL)AMINO]PYRIDIN-4-YL}-4-METHOXYPYRIMIDINE-5-CARBOXAMIDE) to GSK3b | | Descriptor: | 2-{2-[(cyclopropylcarbonyl)amino]pyridin-4-yl}-4-methoxypyrimidine-5-carboxamide, Glycogen synthase kinase-3 beta | | Authors: | Lewis, H.A, Sivaprakasam, P, Kish, K, Pokross, M, Dubowchik, G.M. | | Deposit date: | 2014-03-10 | | Release date: | 2015-04-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.361 Å) | | Cite: | Discovery of new acylaminopyridines as GSK-3 inhibitors by a structure guided in-depth exploration of chemical space around a pyrrolopyridinone core.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
7TQ8
 
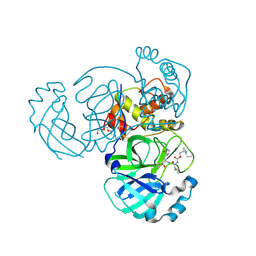 | | Structure of MERS 3CL protease in complex with the cyclopropane based inhibitor 14d | | Descriptor: | (1R,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-{[N-({[(1R,2S)-2-propylcyclopropyl]methoxy}carbonyl)-L-leucyl]amino}propane-1-sulfonic acid, (1S,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-{[N-({[(1R,2S)-2-propylcyclopropyl]methoxy}carbonyl)-L-leucyl]amino}propane-1-sulfonic acid, Orf1a protein, ... | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-01-26 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
7TQ2
 
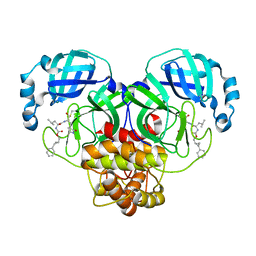 | | Structure of SARS-CoV-2 3CL protease in complex with the cyclopropane based inhibitor 1c | | Descriptor: | 3C-like proteinase, N-{(2S)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-N~2~-({[(1R,2R)-2-phenylcyclopropyl]methoxy}carbonyl)-L-leucinamide | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-01-26 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
