6XS9
 
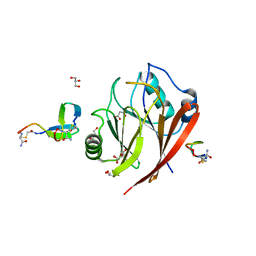 | | Crystal structure of human Vps29 complexed with RaPID-derived cyclic peptide RT-L1 | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, 48V-TYR-ILE-LYS-THR-PRO-LEU-GLY-THR-PHE-PRO-ASN-ARG-HIS-GLY, GLYCEROL, ... | | Authors: | Chen, K.-E, Guo, Q, Collins, B.M. | | Deposit date: | 2020-07-15 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | De novo macrocyclic peptides for inhibiting, stabilizing, and probing the function of the retromer endosomal trafficking complex.
Sci Adv, 7, 2021
|
|
2WBY
 
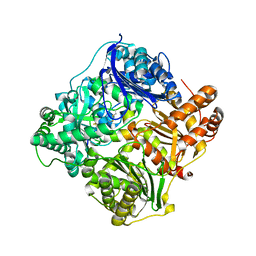 | | Crystal structure of human insulin-degrading enzyme in complex with insulin | | Descriptor: | INSULIN A CHAIN, INSULIN B CHAIN, INSULIN-DEGRADING ENZYME, ... | | Authors: | Manolopoulou, M, Guo, Q, Malito, E, Schilling, A.B, Tang, W.J. | | Deposit date: | 2009-03-06 | | Release date: | 2009-03-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular Basis of Catalytic Chamber-Assisted Unfolding and Cleavage of Human Insulin by Human Insulin Degrading Enzyme.
J.Biol.Chem., 284, 2009
|
|
4CSU
 
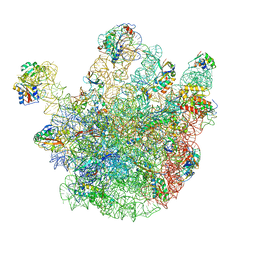 | | Cryo-EM structures of the 50S ribosome subunit bound with ObgE | | Descriptor: | 23S RRNA, 50S RIBOSOMAL PROTEIN L1, 50S RIBOSOMAL PROTEIN L11, ... | | Authors: | Feng, B, Mandava, C.S, Guo, Q, Wang, J, Cao, W, Li, N, Zhang, Y, Zhang, Y, Wang, Z, Wu, J, Sanyal, S, Lei, J, Gao, N. | | Deposit date: | 2014-03-10 | | Release date: | 2014-06-04 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Structural and Functional Insights Into the Mode of Action of a Universally Conserved Obg Gtpase.
Plos Biol., 12, 2014
|
|
1Y0V
 
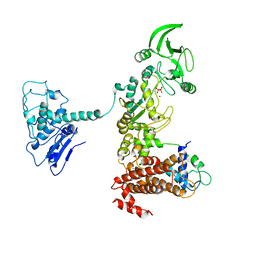 | | Crystal structure of anthrax edema factor (EF) in complex with calmodulin and pyrophosphate | | Descriptor: | CALCIUM ION, Calmodulin, Calmodulin-sensitive adenylate cyclase, ... | | Authors: | Shen, Y, Zhukovskaya, N.L, Guo, Q, Florian, J, Tang, W.-J. | | Deposit date: | 2004-11-16 | | Release date: | 2005-05-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Calcium-independent calmodulin binding and two-metal-ion catalytic mechanism of anthrax edema factor
Embo J., 24, 2005
|
|
6XS7
 
 | | Crystal structure of human Vps29 complexed with RaPID-derived cyclic peptide RT-D2 | | Descriptor: | 48V-DTY-THR-THR-ILE-TYR-TRP-THR-PRO-LEU-GLY-THR-PHE-PRO-ARG-ILE-ARG, FORMIC ACID, GLYCEROL, ... | | Authors: | Chen, K.-E, Guo, Q, Collins, B.M. | | Deposit date: | 2020-07-15 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | De novo macrocyclic peptides for inhibiting, stabilizing, and probing the function of the retromer endosomal trafficking complex.
Sci Adv, 7, 2021
|
|
6XSA
 
 | | Crystal structure of human Vps29 complexed with RaPID-derived cyclic peptide RT-L2 | | Descriptor: | 48V-TYR-LEU-PRO-THR-ILE-THR-GLY-VAL-GLY-HIS-LEU-TRP-HIS-PRO-LEU, SULFATE ION, Vacuolar protein sorting-associated protein 29 | | Authors: | Chen, K.-E, Guo, Q, Collins, B.M. | | Deposit date: | 2020-07-15 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | De novo macrocyclic peptides for inhibiting, stabilizing, and probing the function of the retromer endosomal trafficking complex.
Sci Adv, 7, 2021
|
|
6XS5
 
 | | Crystal structure of human Vps29 complexed with RaPID-derived cyclic peptide RT-D1 | | Descriptor: | 48V-DTY-ILE-ILE-ASP-THR-PRO-LEU-GLY-VAL-PHE-LEU-SER-SER-LEU-LYS-ARG, FORMIC ACID, GLYCEROL, ... | | Authors: | Chen, K.-E, Guo, Q, Collins, B.M. | | Deposit date: | 2020-07-15 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | De novo macrocyclic peptides for inhibiting, stabilizing, and probing the function of the retromer endosomal trafficking complex.
Sci Adv, 7, 2021
|
|
6XS8
 
 | | Crystal structure of Chaetomium thermophilum Vps29 complexed with RaPID-derived cyclic peptide RT-D3 | | Descriptor: | 48V-DTY-GLY-TYR-ASP-PRO-LEU-GLY-LEU-LYS-TYR-PHE-ALA, Vacuolar protein sorting-associated protein 29 | | Authors: | Chen, K.-E, Guo, Q, Collins, B.M. | | Deposit date: | 2020-07-15 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95009851 Å) | | Cite: | De novo macrocyclic peptides for inhibiting, stabilizing, and probing the function of the retromer endosomal trafficking complex.
Sci Adv, 7, 2021
|
|
2PQ3
 
 | | N-Terminal Calmodulin Zn-Trapped Intermediate | | Descriptor: | CACODYLATE ION, Calmodulin, ZINC ION | | Authors: | Warren, J.T, Guo, Q, Tang, W.J. | | Deposit date: | 2007-05-01 | | Release date: | 2007-10-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | A 1.3-A structure of zinc-bound N-terminal domain of calmodulin elucidates potential early ion-binding step.
J.Mol.Biol., 374, 2007
|
|
1XUO
 
 | | X-ray structure of LFA-1 I-domain bound to a 1,4-diazepane-2,5-dione inhibitor at 1.8A resolution | | Descriptor: | (2R)-2-[3-ISOBUTYL-2,5-DIOXO-4-(QUINOLIN-3-YLMETHYL)-1,4-DIAZEPAN-1-YL]-N-METHYL-3-(2-NAPHTHYL)PROPANAMIDE, Integrin alpha-L, MAGNESIUM ION | | Authors: | Wattanasin, S, Kallen, J, Myers, S, Guo, Q, Sabio, M, Ehrhardt, C, Albert, R, Hommel, U, Weckbecker, G, Welzenbach, K. | | Deposit date: | 2004-10-26 | | Release date: | 2005-10-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1,4-Diazepane-2,5-diones as novel inhibitors of LFA-1
Bioorg.Med.Chem.Lett., 15, 2005
|
|
3N56
 
 | |
3N57
 
 | |
3J3W
 
 | | Atomic model of the immature 50S subunit from Bacillus subtilis (state II-a) | | Descriptor: | 50S ribosomal protein L1, 50S ribosomal protein L11, 50S ribosomal protein L13, ... | | Authors: | Li, N, Guo, Q, Zhang, Y, Yuan, Y, Ma, C, Lei, J, Gao, N. | | Deposit date: | 2013-04-28 | | Release date: | 2013-06-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (10.7 Å) | | Cite: | Cryo-EM structures of the late-stage assembly intermediates of the bacterial 50S ribosomal subunit
Nucleic Acids Res., 41, 2013
|
|
3J3V
 
 | | Atomic model of the immature 50S subunit from Bacillus subtilis (state I-a) | | Descriptor: | 50S ribosomal protein L1, 50S ribosomal protein L11, 50S ribosomal protein L13, ... | | Authors: | Li, N, Guo, Q, Zhang, Y, Yuan, Y, Ma, C, Lei, J, Gao, N. | | Deposit date: | 2013-04-28 | | Release date: | 2013-06-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (13.3 Å) | | Cite: | Cryo-EM structures of the late-stage assembly intermediates of the bacterial 50S ribosomal subunit
Nucleic Acids Res., 41, 2013
|
|
9BRQ
 
 | | Intact V-ATPase State 3 and synaptophysin complex in mouse brain isolated synaptic vesicles | | Descriptor: | Renin receptor cytoplasmic fragment, Ribonuclease kappa, Synaptophysin, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-11 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 631, 2024
|
|
9BRU
 
 | | Intact V-ATPase State 1 in synaptophysin knock-out isolated synaptic vesicles | | Descriptor: | Renin receptor cytoplasmic fragment, Ribonuclease kappa, V-type proton ATPase 116 kDa subunit a 1, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-11 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 631, 2024
|
|
9BRT
 
 | | Intact V-ATPase State 1 and synaptophysin complex in mouse brain isolated synaptic vesicles | | Descriptor: | Renin receptor cytoplasmic fragment, Ribonuclease kappa, Synaptophysin, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-11 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 631, 2024
|
|
9BRS
 
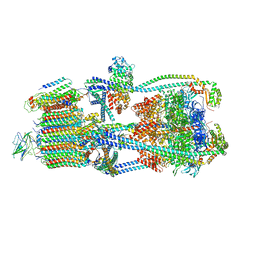 | | Intact V-ATPase State 2 in synaptophysin knock-out isolated synaptic vesicles | | Descriptor: | Renin receptor cytoplasmic fragment, Ribonuclease kappa, V-type proton ATPase 116 kDa subunit a 1, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-11 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 631, 2024
|
|
9BRA
 
 | | Intact V-ATPase State 2 and synaptophysin complex in mouse brain isolated synaptic vesicles | | Descriptor: | Renin receptor cytoplasmic fragment, Ribonuclease kappa, Synaptophysin, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-11 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 631, 2024
|
|
9BRR
 
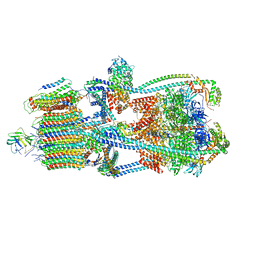 | | Intact V-ATPase State 3 in synaptophysin knock-out isolated synaptic vesicles | | Descriptor: | Renin receptor cytoplasmic fragment, Ribonuclease kappa, V-type proton ATPase 116 kDa subunit a 1, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-11 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 631, 2024
|
|
3BM4
 
 | | Crystal Structure of Human ADP-ribose Pyrophosphatase NUDT5 In complex with magnesium and AMPcpr | | Descriptor: | ADP-sugar pyrophosphatase, ALPHA-BETA METHYLENE ADP-RIBOSE, MAGNESIUM ION | | Authors: | Zha, M, Guo, Q, Zhang, Y, Zhong, C, Ou, Y, Ding, J. | | Deposit date: | 2007-12-12 | | Release date: | 2008-05-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular Mechanism of ADP-Ribose Hydrolysis By Human NUDT5 From Structural and Kinetic Studies
J.Mol.Biol., 379, 2008
|
|
9BRZ
 
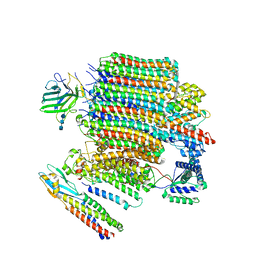 | | V0-only V-ATPase and synaptophysin complex in mouse brain isolated synaptic vesicles | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-12 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 631, 2024
|
|
3H44
 
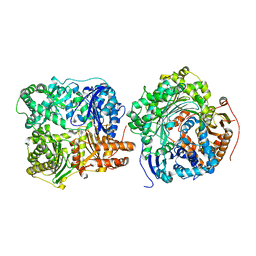 | |
8X5X
 
 | |
8X64
 
 | |
