1ST0
 
 | | Structure of DcpS bound to m7GpppG | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, YTTRIUM (III) ION, mRNA decapping enzyme | | Authors: | Gu, M, Fabrega, C, Liu, S.W, Liu, H, Kiledjian, M, Lima, C.D. | | Deposit date: | 2004-03-24 | | Release date: | 2004-04-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights into the structure, mechanism, and regulation of scavenger mRNA decapping activity
Mol.Cell, 14, 2004
|
|
1ST4
 
 | | Structure of DcpS bound to m7GpppA | | Descriptor: | P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, YTTRIUM (III) ION, mRNA decapping enzyme | | Authors: | Gu, M, Fabrega, C, Liu, S.W, Liu, H, Kiledjian, M, Lima, C.D. | | Deposit date: | 2004-03-24 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Insights into the structure, mechanism, and regulation of scavenger mRNA decapping activity
Mol.Cell, 14, 2004
|
|
3KQH
 
 | |
3KQL
 
 | |
3KQK
 
 | |
3KQN
 
 | |
3KQU
 
 | |
1NI5
 
 | |
5E4F
 
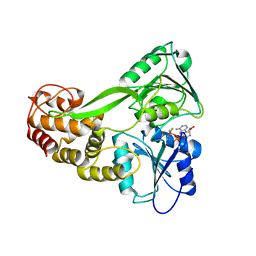 | |
4DYU
 
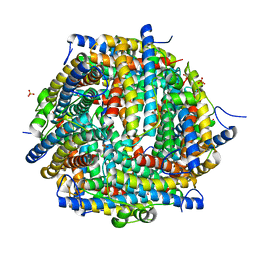 | | The crystal structure of DNA starvation/stationary phase protection protein Dps from Yersinia pestis KIM 10 | | Descriptor: | DNA protection during starvation protein, SULFATE ION, ZINC ION | | Authors: | Tan, K, Gu, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-02-29 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The crystal structure of DNA starvation/stationary phase protection protein Dps from Yersinia pestis KIM 10
To be Published
|
|
4E6Y
 
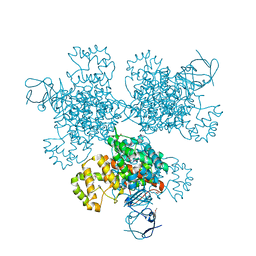 | | Type II citrate synthase from Vibrio vulnificus. | | Descriptor: | Citrate synthase, FORMIC ACID | | Authors: | Osipiuk, J, Gu, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-16 | | Release date: | 2012-03-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Type II citrate synthase from Vibrio vulnificus.
To be Published
|
|
3OZH
 
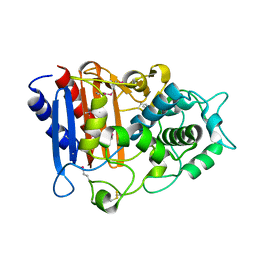 | | Crystal Structure of Beta-Lactamase/D-alanine Carboxypeptidase from Yersinia pestis | | Descriptor: | beta-lactamase/D-alanine carboxypeptidase | | Authors: | Kim, Y, Zhou, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-24 | | Release date: | 2010-10-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.907 Å) | | Cite: | Crystal Structure of Beta-Lactamase/D-alanine Carboxypeptidase from Yersinia pestis
To be Published
|
|
4MY8
 
 | | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase with an Internal Deletion of the CBS Domain from Bacillus anthracis str. Ames complexed with inhibitor Q21 | | Descriptor: | (2S)-2-(naphthalen-1-yloxy)-N-[2-(pyridin-4-yl)-1,3-benzoxazol-5-yl]propanamide, 1,2-ETHANEDIOL, ACETIC ACID, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Gorla, S.K, Kavitha, M, Cuny, G, Hedstrom, L, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-27 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2924 Å) | | Cite: | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase with an Internal Deletion of the CBS Domain from Bacillus anthracis str. Ames complexed with inhibitor Q21
To be Published, 2013
|
|
3OOP
 
 | | The structure of a protein with unknown function from Listeria innocua Clip11262 | | Descriptor: | Lin2960 protein | | Authors: | Fan, Y, Li, H, Zhou, Y, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-31 | | Release date: | 2010-09-22 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The structure of a protein with unknown function from Listeria innocua Clip11262
To be Published
|
|
3OP9
 
 | |
5JMU
 
 | | The crystal structure of the catalytic domain of peptidoglycan N-acetylglucosamine deacetylase from Eubacterium rectale ATCC 33656 | | Descriptor: | ACETATE ION, MAGNESIUM ION, Peptidoglycan N-acetylglucosamine deacetylase, ... | | Authors: | Tan, K, Gu, M, Clancy, S, Joachimiak, A. | | Deposit date: | 2016-04-29 | | Release date: | 2016-06-29 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | The crystal structure of the catalytic domain of peptidoglycan N-acetylglucosamine deacetylase from Eubacterium rectale ATCC 33656 (CASP target)
To Be Published
|
|
4MJM
 
 | | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with a Short Internal Deletion of CBS Domain from Bacillus anthracis str. Ames | | Descriptor: | 1,2-ETHANEDIOL, Inosine-5'-monophosphate dehydrogenase | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-03 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2544 Å) | | Cite: | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with a Short Internal Deletion of CBS Domain from Bacillus anthracis str. Ames
To be Published, 2013
|
|
4MYA
 
 | | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase with an Internal Deletion of the CBS Domain from Bacillus anthracis str. Ames complexed with inhibitor A110 | | Descriptor: | 4-{(1R)-1-[1-(4-chlorophenyl)-1H-1,2,3-triazol-4-yl]ethoxy}quinolin-2(1H)-one, GLYCEROL, INOSINIC ACID, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Gorla, S.K, Hedstrom, L, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-27 | | Release date: | 2014-01-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8997 Å) | | Cite: | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase with an Internal Deletion of the CBS Domain from Bacillus anthracis str. Ames complexed with inhibitor A110
To be Published
|
|
4MYX
 
 | | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with a Internal Deletion of CBS Domain from Bacillus anthracis str. Ame complexed with P32 | | Descriptor: | 1,2-ETHANEDIOL, 2-chloro-5-{[(2-{3-[(1E)-N-hydroxyethanimidoyl]phenyl}propan-2-yl)carbamoyl]amino}benzamide, FORMIC ACID, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Gorla, S.K, Hedstrom, L, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-28 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with a Internal Deletion of CBS Domain from Bacillus anthracis str. Ame complexed with P32
To be Published
|
|
4MY9
 
 | | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase with an Internal Deletion of the CBS Domain from Bacillus anthracis str. Ames complexed with inhibitor C91 | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, MALONATE ION, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Gorla, S.K, Hedstrom, L, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-27 | | Release date: | 2014-06-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5893 Å) | | Cite: | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase with an Internal Deletion of the CBS Domain from Bacillus anthracis str. Ames complexed with inhibitor C91
To be Published
|
|
4MY1
 
 | | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with a Internal Deletion of CBS Domain from Bacillus anthracis str. Ames complexed with P68 | | Descriptor: | 1-(4-bromophenyl)-3-(2-{3-[(1E)-N-hydroxyethanimidoyl]phenyl}propan-2-yl)urea, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Gorla, S.K, Hedstrom, L, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-26 | | Release date: | 2014-01-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5997 Å) | | Cite: | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with a Internal Deletion of CBS Domain from Bacillus anthracis str. Ames complexed with P68
To be Published
|
|
4MZ8
 
 | | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with an Internal Deletion of CBS Domain from Campylobacter jejuni complexed with inhibitor compound C91 | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Gorla, S.K, Hedstrom, L, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-29 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5004 Å) | | Cite: | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with a Internal Deletion of CBS Domain from Campylobacter jejuni complexed with inhibitor compound C91
To be Published
|
|
9CS1
 
 | | E. coli BamA beta-barrel bound to darobactin and cyclic peptide CP2 | | Descriptor: | Cyclic peptide CP2, Darobactin A, Outer membrane protein assembly factor BamA, ... | | Authors: | Walker, M.E, Gu, M, Lu, J, Klein, D.J. | | Deposit date: | 2024-07-23 | | Release date: | 2025-04-16 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2.147 Å) | | Cite: | Antibacterial macrocyclic peptides reveal a distinct mode of BamA inhibition.
Nat Commun, 16, 2025
|
|
9CS0
 
 | | E. coli BamA beta-barrel bound to darobactin and cyclic peptide CP1 | | Descriptor: | Cyclic peptide CP1, Darobactin A, Outer membrane protein assembly factor BamA, ... | | Authors: | Walker, M.E, Gu, M, Lu, J, Klein, D.J. | | Deposit date: | 2024-07-23 | | Release date: | 2025-04-16 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.944 Å) | | Cite: | Antibacterial macrocyclic peptides reveal a distinct mode of BamA inhibition.
Nat Commun, 16, 2025
|
|
9CS2
 
 | | E. coli BamA beta-barrel bound to cyclic peptide CP3 | | Descriptor: | Cyclic peptide CP3, Outer membrane protein assembly factor BamA, tetrabutylphosphonium | | Authors: | Walker, M.E, Gu, M, Lu, J, Klein, D.J. | | Deposit date: | 2024-07-23 | | Release date: | 2025-04-16 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.784 Å) | | Cite: | Antibacterial macrocyclic peptides reveal a distinct mode of BamA inhibition.
Nat Commun, 16, 2025
|
|
