7Z28
 
 | | High-resolution crystal structure of ERAP1 with bound bestatin analogue inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Giastas, P, Papakyriakou, A, Stratikos, E, Vourloumis, D. | | Deposit date: | 2022-02-26 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of Selective Nanomolar Inhibitors for Insulin-Regulated Aminopeptidase Based on alpha-Hydroxy-beta-amino Acid Derivatives of Bestatin.
J.Med.Chem., 65, 2022
|
|
6Q4R
 
 | | High-resolution crystal structure of ERAP1 with bound phosphinic transition-state analogue inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Giastas, P, Neu, M, Rowland, P, Stratikos, E. | | Deposit date: | 2018-12-06 | | Release date: | 2019-04-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-Resolution Crystal Structure of Endoplasmic Reticulum Aminopeptidase 1 with Bound Phosphinic Transition-State Analogue Inhibitor.
Acs Med.Chem.Lett., 10, 2019
|
|
4UY2
 
 | | Crystal structure of the complex of the extracellular domain of human alpha9 nAChR with alpha-bungarotoxin. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-BUNGAROTOXIN ISOFORM V31, NEURONAL ACETYLCHOLINE RECEPTOR SUBUNIT ALPHA-9 | | Authors: | Giastas, P, Zouridakis, M, Zarkadas, E, Tzartos, S.J. | | Deposit date: | 2014-08-28 | | Release date: | 2014-10-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.697 Å) | | Cite: | Crystal Structures of Free and Antagonist-Bound States of Human Alpha9 Nicotinic Receptor Extracellular Domain
Nat.Struct.Mol.Biol., 21, 2014
|
|
4V33
 
 | | Crystal structure of the putative polysaccharide deacetylase BA0330 from bacillus anthracis | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, POLYSACCHARIDE DEACETYLASE-LIKE PROTEIN, ... | | Authors: | Giastas, P, Andreou, A, Bouriotis, V, Eliopoulos, E. | | Deposit date: | 2014-10-16 | | Release date: | 2015-04-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Two Putative Polysaccharide Deacetylases are Required for Osmotic Stability and Cell Shape Maintenance in Bacillus Anthracis.
J.Biol.Chem., 290, 2015
|
|
3EXY
 
 | |
2ZVS
 
 | | Crystal structure of the 2[4FE-4S] ferredoxin from escherichia coli | | Descriptor: | IRON/SULFUR CLUSTER, Uncharacterized ferredoxin-like protein yfhL | | Authors: | Giastas, P, Mavridis, M.I. | | Deposit date: | 2008-11-18 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Insight into the protein and solvent contributions to the reduction potentials of [4Fe-4S]2+/+ clusters: crystal structures of the Allochromatium vinosum ferredoxin variants C57A and V13G and the homologous Escherichia coli ferredoxin
J.Biol.Inorg.Chem., 14, 2009
|
|
2FGO
 
 | | Structure of the 2[4FE-4S] ferredoxin from Pseudomonas aeruginosa | | Descriptor: | Ferredoxin, IRON/SULFUR CLUSTER | | Authors: | Giastas, P, Pinotsis, N, Mavridis, I.M. | | Deposit date: | 2005-12-22 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | The structure of the 2[4Fe-4S] ferredoxin from Pseudomonas aeruginosa at 1.32-A resolution: comparison with other high-resolution structures of ferredoxins and contributing structural features to reduction potential values.
J.Biol.Inorg.Chem., 11, 2006
|
|
4D01
 
 | | Crystal Structure of the Extracellular Domain of the Human Alpha9 Nicotinic Acetylcholine Receptor | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, NEURONAL ACETYLCHOLINE RECEPTOR SUBUNIT ALPHA-9 | | Authors: | Giastas, P, Zouridakis, M, Zarkadas, E, Tzartos, S.J. | | Deposit date: | 2014-04-23 | | Release date: | 2014-10-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.795 Å) | | Cite: | Crystal Structures of Free and Antagonist-Bound States of Human Alpha9 Nicotinic Receptor Extracellular Domain
Nat.Struct.Mol.Biol., 21, 2014
|
|
6GO1
 
 | | Crystal Structure of a Bacillus anthracis peptidoglycan deacetylase | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Polysaccharide deacetylase-like protein, ... | | Authors: | Giastas, P, Andreou, A, Eliopoulos, E.E. | | Deposit date: | 2018-06-01 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | The putative polysaccharide deacetylase Ba0331: cloning, expression, crystallization and structure determination.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
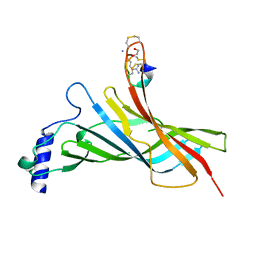
6HY7
 
 | |
6RQX
 
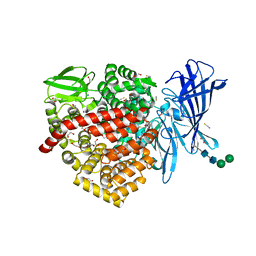 | |
8A5U
 
 | |
6RYF
 
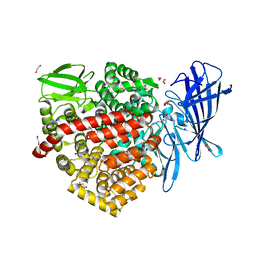 | | High-resolution crystal structure of ERAP1 in complex with 15mer phosphinic peptide | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Giastas, P, Stratikos, E. | | Deposit date: | 2019-06-10 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Mechanism for antigenic peptide selection by endoplasmic reticulum aminopeptidase 1.
Proc.Natl.Acad.Sci.USA, 2019
|
|
5FJV
 
 | | Crystal structure of the extracellular domain of alpha2 nicotinic acetylcholine receptor in pentameric assembly | | Descriptor: | EPIBATIDINE, NEURONAL ACETYLCHOLINE RECEPTOR SUBUNIT ALPHA-2 | | Authors: | Giastas, P, Kouvatsos, N, Chroni-Tzartou, D, Tzartos, S.J. | | Deposit date: | 2015-10-13 | | Release date: | 2016-08-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of a Human Neuronal Nachr Extracellular Domain in Pentameric Assembly: Ligand-Bound Alpah2 Homopentamer.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7P7P
 
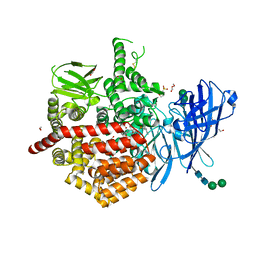 | | Crystal structure of ERAP2 aminopeptidase in complex with phosphinic pseudotripeptide((1R)-1-Amino-3-phenylpropyl){(2S)-3-[((2S)-1-amino-1-oxo-3-phenylpropan-2-yl)amino]-2-{[3-(2-hydroxyphenyl)-isoxazol-5-yl]methyl}-3-oxopropyl}phosphinic acid | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Giastas, P, Stratikos, E, Mpakali, A. | | Deposit date: | 2021-07-20 | | Release date: | 2022-01-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Inhibitor-Dependent Usage of the S1' Specificity Pocket of ER Aminopeptidase 2.
Acs Med.Chem.Lett., 13, 2022
|
|
7PFS
 
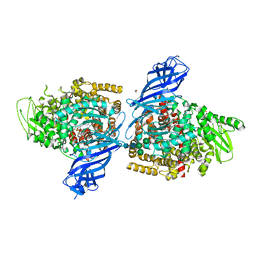 | | Crystal structure of ERAP2 aminopeptidase in complex with phosphinic pseudotripeptide ((1R)-1-Amino-3-phenylpropyl){2-([1,1:3,1-terphenyl]-5-ylmethyl)-3-[((2S)-1-amino-1-oxo-3-phenylpropan-2-yl)-amino]-3-oxopropyl}phosphinic acid | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Giastas, P, Stratikos, E, Mpakali, A. | | Deposit date: | 2021-08-12 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Inhibitor-Dependent Usage of the S1' Specificity Pocket of ER Aminopeptidase 2.
Acs Med.Chem.Lett., 13, 2022
|
|
5N1J
 
 | | Crystal structure of the polysaccharide deacetylase Bc1974 from Bacillus cereus | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Peptidoglycan N-acetylglucosamine deacetylase, ... | | Authors: | Giastas, P, Andreou, A, Balomenou, S, Bouriotis, V, Eliopoulos, E.E. | | Deposit date: | 2017-02-06 | | Release date: | 2018-02-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of the Peptidoglycan N-Acetylglucosamine Deacetylase Bc1974 and Its Complexes with Zinc Metalloenzyme Inhibitors.
Biochemistry, 57, 2018
|
|
5N1P
 
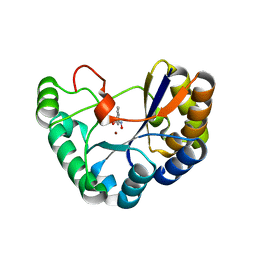 | | Crystal structure of the polysaccharide deacetylase Bc1974 from Bacillus cereus in complex with N-hydroxynaphthalene-1-carboxamide | | Descriptor: | 1,2-ETHANEDIOL, Peptidoglycan N-acetylglucosamine deacetylase, SODIUM ION, ... | | Authors: | Giastas, P, Andreou, A, Eliopoulos, E.E. | | Deposit date: | 2017-02-06 | | Release date: | 2018-02-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.448 Å) | | Cite: | Structures of the Peptidoglycan N-Acetylglucosamine Deacetylase Bc1974 and Its Complexes with Zinc Metalloenzyme Inhibitors.
Biochemistry, 57, 2018
|
|
5NCD
 
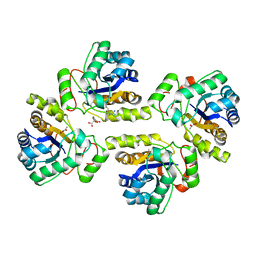 | | Crystal structure of the polysaccharide deacetylase Bc1974 from Bacillus cereus in complex with (2S)-2-amino-5-(diaminomethylideneamino)-N-hydroxypentanamide | | Descriptor: | ACETATE ION, CITRIC ACID, N-HYDROXY-L-ARGININAMIDE, ... | | Authors: | Giastas, P, Andreou, A, Eliopoulos, E.E. | | Deposit date: | 2017-03-03 | | Release date: | 2018-02-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.447 Å) | | Cite: | Structures of the Peptidoglycan N-Acetylglucosamine Deacetylase Bc1974 and Its Complexes with Zinc Metalloenzyme Inhibitors.
Biochemistry, 57, 2018
|
|
5NC6
 
 | | Crystal structure of the polysaccharide deacetylase Bc1974 from Bacillus cereus in complex with (E)-N-hydroxy-3-(naphthalen-1-yl)prop-2-enamide | | Descriptor: | 1,2-ETHANEDIOL, 3-naphthalen-1-yl-~{N}-oxidanyl-propanamide, ACETATE ION, ... | | Authors: | Giastas, P, Andreou, A, Balomenou, S, Bouriotis, V, Eliopoulos, E.E. | | Deposit date: | 2017-03-03 | | Release date: | 2018-02-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of the Peptidoglycan N-Acetylglucosamine Deacetylase Bc1974 and Its Complexes with Zinc Metalloenzyme Inhibitors.
Biochemistry, 57, 2018
|
|
5NC9
 
 | | Crystal structure of the polysaccharide deacetylase Bc1974 from Bacillus cereus in complex with (2S)-2,6-diamino-N-hydroxyhexanamide | | Descriptor: | (2~{S})-2,6-bis(azanyl)-~{N}-oxidanyl-hexanamide, 1,2-ETHANEDIOL, CITRIC ACID, ... | | Authors: | Giastas, P, Andreou, A, Eliopoulos, E.E. | | Deposit date: | 2017-03-03 | | Release date: | 2018-02-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structures of the Peptidoglycan N-Acetylglucosamine Deacetylase Bc1974 and Its Complexes with Zinc Metalloenzyme Inhibitors.
Biochemistry, 57, 2018
|
|
4UXU
 
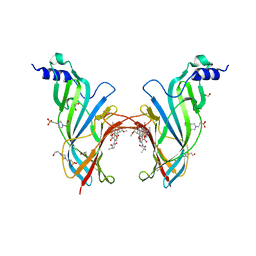 | | Crystal Structure of the Extracellular Domain of the Human Alpha9 Nicotinic Acetylcholine Receptor In Complex with Methyllycaconitine | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Zouridakis, M, Giastas, P, Zarkadas, E, Tzartos, S.J. | | Deposit date: | 2014-08-27 | | Release date: | 2014-10-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal Structures of Free and Antagonist-Bound States of Human Alpha9 Nicotinic Receptor Extracellular Domain
Nat.Struct.Mol.Biol., 21, 2014
|
|
8P0I
 
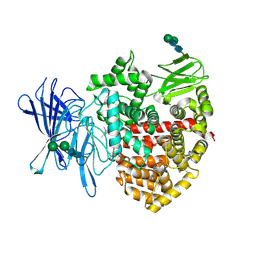 | | Crystal structure of the open conformation of insulin-regulated aminopeptidase in complex with a small-MW inhibitor | | Descriptor: | 2-[2-(3,5-dimethylpyrazol-1-yl)-6-(4-methoxyphenyl)pyrimidin-4-yl]oxy-N-methyl-N-[(2-methylpyridin-3-yl)methyl]ethanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mpakali, A, Giastas, P, Stratikos, E. | | Deposit date: | 2023-05-10 | | Release date: | 2024-09-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Stabilization of the open conformation omicron f insulin-regulated aminopeptidase by a novel substrate-selective small-molecule inhibitor.
Protein Sci., 33, 2024
|
|
5K1V
 
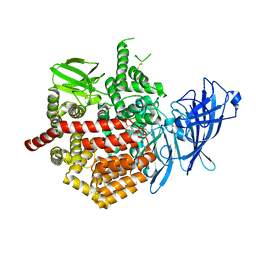 | | Crystal structure of Endoplasmic Reticulum aminopeptidase 2 (ERAP2) in complex with a diaminobenzoic acid derivative ligand. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Endoplasmic reticulum aminopeptidase 2, ... | | Authors: | Saridakis, E, Papakyriakou, A, Giastas, P, Mpakali, A, Mavridis, I.M, Stratikos, E. | | Deposit date: | 2016-05-18 | | Release date: | 2017-03-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.897 Å) | | Cite: | Crystal Structures of ERAP2 Complexed with Inhibitors Reveal Pharmacophore Requirements for Optimizing Inhibitor Potency.
ACS Med Chem Lett, 8, 2017
|
|
5CU5
 
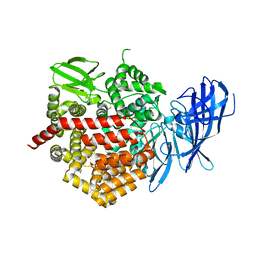 | | Crystal structure of ERAP2 without catalytic Zn(II) atom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Endoplasmic reticulum aminopeptidase 2, ... | | Authors: | Saridakis, E, Mathioudakis, N, Giastas, P, Mavridis, I.M, Stratikos, E. | | Deposit date: | 2015-07-24 | | Release date: | 2015-09-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Structural Basis for Antigenic Peptide Recognition and Processing by Endoplasmic Reticulum (ER) Aminopeptidase 2.
J.Biol.Chem., 290, 2015
|
|
