1QA5
 
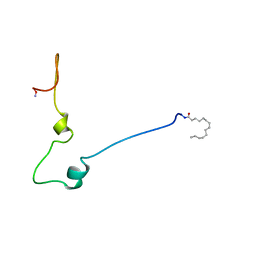 | |
1QA4
 
 | |
2RGF
 
 | | RBD OF RAL GUANOSINE-NUCLEOTIDE EXCHANGE FACTOR (PROTEIN), NMR, 10 STRUCTURES | | Descriptor: | RALGEF-RBD | | Authors: | Geyer, M, Herrmann, C, Wittinghofer, A, Kalbitzer, H.R. | | Deposit date: | 1997-02-13 | | Release date: | 1998-03-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the Ras-binding domain of RalGEF and implications for Ras binding and signalling.
Nat.Struct.Biol., 4, 1997
|
|
4ORZ
 
 | |
9FMR
 
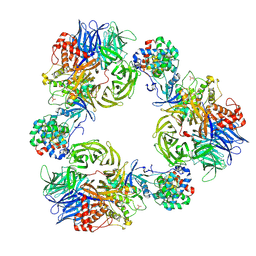 | | Structure of DDB1/Cdk12/Cyclin K with molecular glue SR-4835 | | Descriptor: | Cyclin-K, Cyclin-dependent kinase 12, DNA damage-binding protein 1, ... | | Authors: | Anand, K, Schmitz, M, Geyer, M. | | Deposit date: | 2024-06-07 | | Release date: | 2024-10-16 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Discovery and design of molecular glue enhancers of CDK12-DDB1 interactions for targeted degradation of cyclin K.
Rsc Chem Biol, 6, 2024
|
|
8PYR
 
 | | Crystal structure of the dual T-loop phosphorylated Cdk7/CycH/Mat1 complex | | Descriptor: | 1,2-ETHANEDIOL, CDK-activating kinase assembly factor MAT1, Cyclin-H, ... | | Authors: | Anand, K, Duster, R, Geyer, M. | | Deposit date: | 2023-07-26 | | Release date: | 2024-03-06 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis of Cdk7 activation by dual T-loop phosphorylation.
Nat Commun, 15, 2024
|
|
8P81
 
 | | Crystal structure of human Cdk12/Cyclin K in complex with inhibitor SR-4835 | | Descriptor: | Cyclin-K, Cyclin-dependent kinase 12, ~{N}-[[5,6-bis(chloranyl)-1~{H}-benzimidazol-2-yl]methyl]-9-(1-methylpyrazol-4-yl)-2-morpholin-4-yl-purin-6-amine | | Authors: | Anand, K, Schmitz, M, Geyer, M. | | Deposit date: | 2023-05-31 | | Release date: | 2023-11-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | The reversible inhibitor SR-4835 binds Cdk12/cyclin K in a noncanonical G-loop conformation.
J.Biol.Chem., 300, 2023
|
|
8CMU
 
 | | High resolution structure of the coagulation Factor XIII A2B2 heterotetramer complex. | | Descriptor: | Coagulation factor XIII A chain, Coagulation factor XIII B chain | | Authors: | Singh, S, Urgular, D, Hagelueken, G, Geyer, M, Biswas, A. | | Deposit date: | 2023-02-21 | | Release date: | 2024-09-11 | | Last modified: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | Cryo-EM structure of the human native plasma coagulation factor XIII complex.
Blood, 145, 2025
|
|
8CMT
 
 | | Structure of the plasma coagulation Factor XIII A2B2 heterotetrameric complex. | | Descriptor: | Coagulation factor XIII A chain, Coagulation factor XIII B chain | | Authors: | Singh, S, Ugurlar, D, Hagelueken, G, Geyer, M, Biswas, A. | | Deposit date: | 2023-02-21 | | Release date: | 2024-09-11 | | Last modified: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Cryo-EM structure of the human native plasma coagulation factor XIII complex.
Blood, 145, 2025
|
|
7Z1X
 
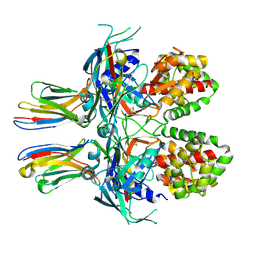 | |
7PZC
 
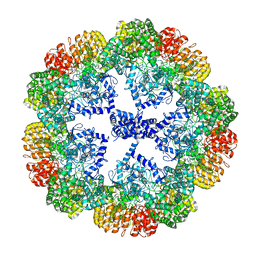 | | Cryo-EM structure of the NLRP3 decamer bound to the inhibitor CRID3 | | Descriptor: | 1-(1,2,3,5,6,7-hexahydro-s-indacen-4-yl)-3-[4-(2-oxidanylpropan-2-yl)furan-2-yl]sulfonyl-urea, ADENOSINE-5'-DIPHOSPHATE, NACHT, ... | | Authors: | Hochheiser, I.V, Pilsl, M, Hagelueken, G, Engel, C, Geyer, M. | | Deposit date: | 2021-10-12 | | Release date: | 2022-01-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the NLRP3 decamer bound to the cytokine release inhibitor CRID3.
Nature, 604, 2022
|
|
6Z2G
 
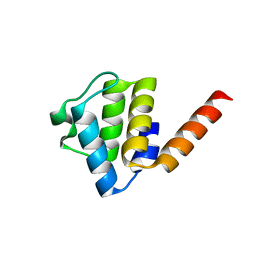 | | Crystal structure of human NLRP9 PYD | | Descriptor: | NACHT, LRR and PYD domains-containing protein 9 | | Authors: | Marleaux, M, Anand, K, Geyer, M. | | Deposit date: | 2020-05-15 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the human NLRP9 pyrin domain suggests a distinct mode of inflammasome assembly.
Febs Lett., 594, 2020
|
|
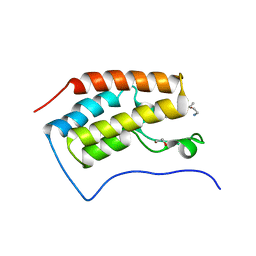
3MUL
 
 | |
3MUK
 
 | |
4NST
 
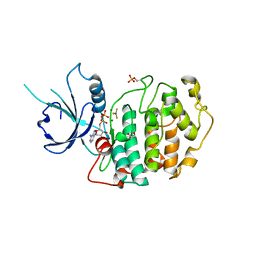 | | Crystal structure of human Cdk12/Cyclin K in complex with ADP-aluminum fluoride | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, ... | | Authors: | Boesken, C.A, Farnung, L, Anand, K, Geyer, M. | | Deposit date: | 2013-11-29 | | Release date: | 2014-03-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure and substrate specificity of human Cdk12/Cyclin K.
Nat Commun, 5, 2014
|
|
5EFQ
 
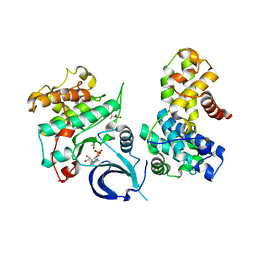 | | Crystal structure of human Cdk13/Cyclin K in complex with ADP-aluminum fluoride | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Cyclin-K, ... | | Authors: | Hoenig, D, Greifenberg, A.K, Anand, K, Geyer, M. | | Deposit date: | 2015-10-24 | | Release date: | 2015-12-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Functional Analysis of the Cdk13/Cyclin K Complex.
Cell Rep, 14, 2016
|
|
9E6M
 
 | | Crystal structure of the G200R mutant from the maize chloroplastic photosynthetic NADP(+)-dependent malic enzyme | | Descriptor: | Malic enzyme, PYRUVIC ACID | | Authors: | Klinke, S, Schneberger, N, Boehm, J.M, Willms, S, Hagelueken, G, Geyer, M, Maurino, V, Alvarez, C.E. | | Deposit date: | 2024-10-30 | | Release date: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the G200R mutant from the maize chloroplastic photosynthetic NADP(+)-dependent malic enzyme
To Be Published
|
|
7PZD
 
 | | Cryo-EM structure of the NLRP3 PYD filament | | Descriptor: | NACHT, LRR and PYD domains-containing protein 3 | | Authors: | Hochheiser, I.V, Hagelueken, G, Behrmann, H, Behrmann, E, Geyer, M. | | Deposit date: | 2021-10-12 | | Release date: | 2022-04-20 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Directionality of PYD filament growth determined by the transition of NLRP3 nucleation seeds to ASC elongation.
Sci Adv, 8, 2022
|
|
3DAD
 
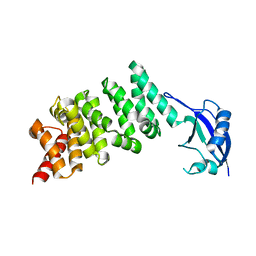 | | Crystal structure of the N-terminal regulatory domains of the formin FHOD1 | | Descriptor: | FH1/FH2 domain-containing protein 1 | | Authors: | Schulte, A, Stolp, B, Schonichen, A, Pylypenko, O, Rak, A, Fackler, O.T, Geyer, M. | | Deposit date: | 2008-05-29 | | Release date: | 2008-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Human Formin FHOD1 Contains a Bipartite Structure of FH3 and GTPase-Binding Domains Required for Activation.
Structure, 16, 2008
|
|
3REB
 
 | |
1G6P
 
 | | SOLUTION NMR STRUCTURE OF THE COLD SHOCK PROTEIN FROM THE HYPERTHERMOPHILIC BACTERIUM THERMOTOGA MARITIMA | | Descriptor: | COLD SHOCK PROTEIN TMCSP | | Authors: | Kremer, W, Schuler, B, Harrieder, S, Geyer, M, Gronwald, W, Welker, C, Jaenicke, R, Kalbitzer, H.R. | | Deposit date: | 2000-11-07 | | Release date: | 2001-11-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the cold-shock protein from the hyperthermophilic bacterium Thermotoga maritima.
Eur.J.Biochem., 268, 2001
|
|
7BAI
 
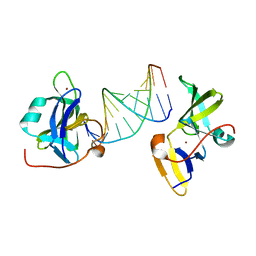 | | Structure of RIG-I CTD (I875A) bound to p-RNA | | Descriptor: | Antiviral innate immune response receptor RIG-I, RNA (5'-R(*(GDP)P*AP*CP*GP*CP*UP*AP*GP*CP*GP*UP*C)-3'), ZINC ION | | Authors: | Anand, K, Hagelueken, G, Fusshoeller, D, Geyer, M. | | Deposit date: | 2020-12-15 | | Release date: | 2022-01-12 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | A conserved isoleucine in the binding pocket of RIG-I controls immune tolerance to mitochondrial RNA.
Nucleic Acids Res., 51, 2023
|
|
7BAH
 
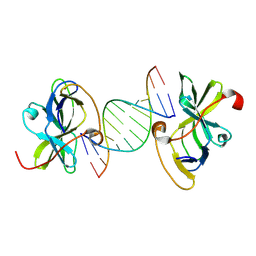 | | Structure of RIG-I CTD bound to OH-RNA | | Descriptor: | Antiviral innate immune response receptor RIG-I, RNA (5'-R(*GP*AP*CP*GP*CP*UP*AP*GP*CP*GP*UP*C)-3'), ZINC ION | | Authors: | Anand, K, Hagelueken, G, Fusshoeller, D, Geyer, M. | | Deposit date: | 2020-12-15 | | Release date: | 2022-01-12 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A conserved isoleucine in the binding pocket of RIG-I controls immune tolerance to mitochondrial RNA.
Nucleic Acids Res., 51, 2023
|
|
2PK2
 
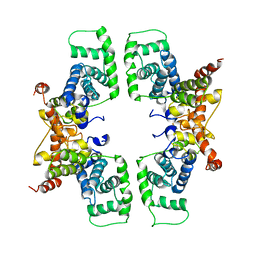 | | Cyclin box structure of the P-TEFb subunit Cyclin T1 derived from a fusion complex with EIAV Tat | | Descriptor: | Cyclin-T1, Protein Tat | | Authors: | Anand, K, Schulte, A, Fujinaga, K, Scheffzek, K, Geyer, M. | | Deposit date: | 2007-04-17 | | Release date: | 2007-07-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Cyclin Box Structure of the P-TEFb Subunit Cyclin T1 Derived from a Fusion Complex with EIAV Tat.
J.Mol.Biol., 370, 2007
|
|
4YDH
 
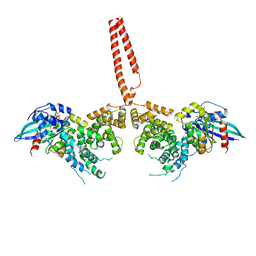 | | The structure of human FMNL1 N-terminal domains bound to Cdc42 | | Descriptor: | Cell division control protein 42 homolog, Formin-like protein 1, MAGNESIUM ION, ... | | Authors: | Kuhn, S, Anand, K, Geyer, M. | | Deposit date: | 2015-02-22 | | Release date: | 2015-05-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | The structure of FMNL2-Cdc42 yields insights into the mechanism of lamellipodia and filopodia formation.
Nat Commun, 6, 2015
|
|
