4MXI
 
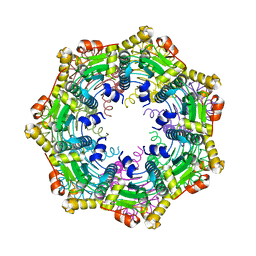 | | ClpP Ser98dhA | | Descriptor: | ATP-dependent Clp protease proteolytic subunit | | Authors: | Gersch, M, Kolb, R, Alte, F, Groll, M, Sieber, S.A. | | Deposit date: | 2013-09-26 | | Release date: | 2013-10-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Disruption of Oligomerization and Dehydroalanine Formation as Mechanisms for ClpP Protease Inhibition.
J.Am.Chem.Soc., 136, 2014
|
|
3V5E
 
 | | Crystal structure of ClpP from Staphylococcus aureus in the active, extended conformation | | Descriptor: | ATP-dependent Clp protease proteolytic subunit | | Authors: | Gersch, M, List, A, Groll, M, Sieber, S. | | Deposit date: | 2011-12-16 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insights into structural network responsible for oligomerization and activity of bacterial virulence regulator caseinolytic protease P (ClpP) protein.
J.Biol.Chem., 287, 2012
|
|
5OHP
 
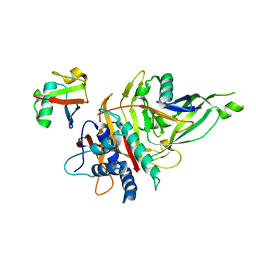 | |
5OHK
 
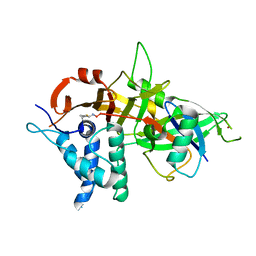 | | Crystal structure of USP30 in covalent complex with ubiquitin propargylamide (high resolution) | | Descriptor: | Polyubiquitin-B, Ubiquitin carboxyl-terminal hydrolase 30,Ubiquitin carboxyl-terminal hydrolase 30,Ubiquitin carboxyl-terminal hydrolase 30, ZINC ION, ... | | Authors: | Gersch, M, Komander, D. | | Deposit date: | 2017-07-17 | | Release date: | 2017-09-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Mechanism and regulation of the Lys6-selective deubiquitinase USP30.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5OHN
 
 | |
6HEJ
 
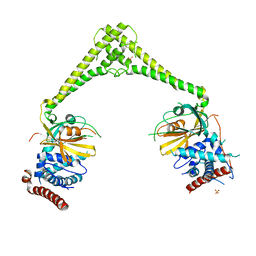 | | Structure of human USP28 | | Descriptor: | SULFATE ION, Ubiquitin carboxyl-terminal hydrolase 28 | | Authors: | Gersch, M, Komander, D. | | Deposit date: | 2018-08-20 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Distinct USP25 and USP28 Oligomerization States Regulate Deubiquitinating Activity.
Mol.Cell, 74, 2019
|
|
6HEM
 
 | |
6HEK
 
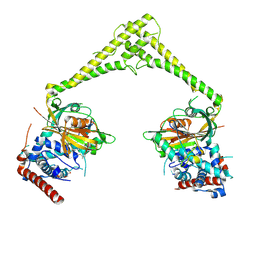 | | Structure of human USP28 bound to Ubiquitin-PA | | Descriptor: | CHLORIDE ION, Polyubiquitin-B, TETRAETHYLENE GLYCOL, ... | | Authors: | Gersch, M, Komander, D. | | Deposit date: | 2018-08-20 | | Release date: | 2019-03-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Distinct USP25 and USP28 Oligomerization States Regulate Deubiquitinating Activity.
Mol.Cell, 74, 2019
|
|
6HEH
 
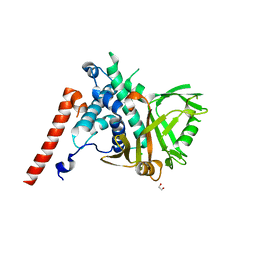 | | Structure of the catalytic domain of USP28 (insertion deleted) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Ubiquitin carboxyl-terminal hydrolase 28,Ubiquitin carboxyl-terminal hydrolase 28 | | Authors: | Gersch, M, Komander, D. | | Deposit date: | 2018-08-20 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Distinct USP25 and USP28 Oligomerization States Regulate Deubiquitinating Activity.
Mol.Cell, 74, 2019
|
|
6HEI
 
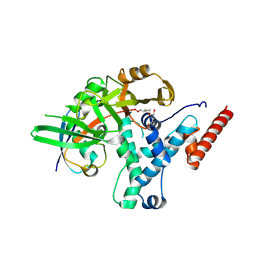 | |
6HEL
 
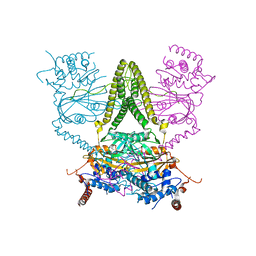 | | Structure of human USP25 | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 25 | | Authors: | Gersch, M, Komander, D. | | Deposit date: | 2018-08-20 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.941 Å) | | Cite: | Distinct USP25 and USP28 Oligomerization States Regulate Deubiquitinating Activity.
Mol.Cell, 74, 2019
|
|
8C61
 
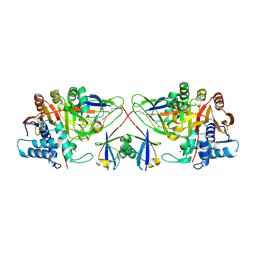 | |
7ZM0
 
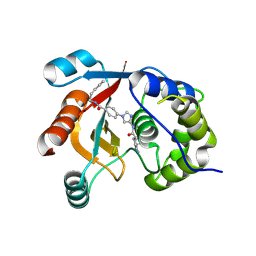 | | Structure of UCHL1 in complex with GK13S inhibitor | | Descriptor: | (3S)-1-(iminomethyl)-N-[1-[4-(pent-4-ynylcarbamoyl)phenyl]imidazol-4-yl]pyrrolidine-3-carboxamide, Ubiquitin carboxyl-terminal hydrolase isozyme L1 | | Authors: | Grethe, C, Gersch, M. | | Deposit date: | 2022-04-18 | | Release date: | 2022-09-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural basis for specific inhibition of the deubiquitinase UCHL1.
Nat Commun, 13, 2022
|
|
9F6G
 
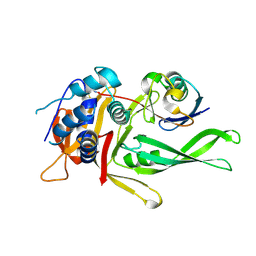 | | Human USP30 chimera bound to Ubiquitin-PA | | Descriptor: | Polyubiquitin-B, Ubiquitin carboxyl-terminal hydrolase 30,Ubiquitin carboxyl-terminal hydrolase 14,Ubiquitin carboxyl-terminal hydrolase 35, prop-2-en-1-amine | | Authors: | Kazi, N.H, Gersch, M. | | Deposit date: | 2024-05-01 | | Release date: | 2025-03-19 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Chimeric deubiquitinase engineering reveals structural basis for specific inhibition of the mitophagy regulator USP30.
Nat.Struct.Mol.Biol., 32, 2025
|
|
9F19
 
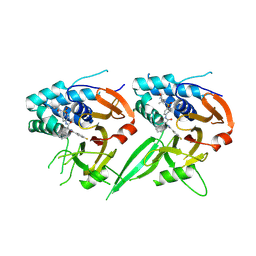 | | Human USP30 chimera in complex with NK036 inhibitor | | Descriptor: | 4-fluoranyl-~{N}-[(2~{S})-1-[[4-[(2-methyl-1-oxidanyl-propan-2-yl)sulfamoyl]phenyl]amino]-1-oxidanylidene-3-phenyl-propan-2-yl]benzamide, Ubiquitin carboxyl-terminal hydrolase 30,Ubiquitin carboxyl-terminal hydrolase 14,Ubiquitin carboxyl-terminal hydrolase 35 | | Authors: | Kazi, N.H, Gersch, M. | | Deposit date: | 2024-04-18 | | Release date: | 2025-03-19 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Chimeric deubiquitinase engineering reveals structural basis for specific inhibition of the mitophagy regulator USP30.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8PW1
 
 | | Structure of human UCHL1 in complex with CG341 inhibitor | | Descriptor: | (2~{S})-4-(iminomethyl)-1-methyl-~{N}-[1-[4-(pent-4-ynylcarbamoyl)phenyl]imidazol-4-yl]piperazine-2-carboxamide, Ubiquitin carboxyl-terminal hydrolase isozyme L1 | | Authors: | Grethe, C, Gersch, M. | | Deposit date: | 2023-07-19 | | Release date: | 2024-01-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | N-Cyanopiperazines as Specific Covalent Inhibitors of the Deubiquitinating Enzyme UCHL1.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8PPW
 
 | | Structure of human PARK7 in complex with GK16S | | Descriptor: | (3~{S})-1-(iminomethyl)-~{N}-pent-4-ynyl-pyrrolidine-3-carboxamide, Parkinson disease protein 7 | | Authors: | Grethe, C, Gersch, M. | | Deposit date: | 2023-07-10 | | Release date: | 2024-01-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | N-Cyanopiperazines as Specific Covalent Inhibitors of the Deubiquitinating Enzyme UCHL1.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8PQ0
 
 | | Structure of human PARK7 in complex with GK16R | | Descriptor: | (3~{R})-3-(pent-4-ynylcarbamoyl)pyrrolidine-1-carboximidothioic acid, Parkinson disease protein 7 | | Authors: | Grethe, C, Gersch, M. | | Deposit date: | 2023-07-10 | | Release date: | 2024-01-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | N-Cyanopiperazines as Specific Covalent Inhibitors of the Deubiquitinating Enzyme UCHL1.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
7ZJV
 
 | |
7ZJU
 
 | |
8BS9
 
 | | Structure of USP36 in complex with Ubiquitin-PA | | Descriptor: | Polyubiquitin-B, SODIUM ION, Ubiquitin carboxyl-terminal hydrolase 36, ... | | Authors: | O'Dea, R, Gersch, M. | | Deposit date: | 2022-11-24 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis for ubiquitin/Fubi cross-reactivity in USP16 and USP36.
Nat.Chem.Biol., 19, 2023
|
|
8BS3
 
 | | Structure of USP36 in complex with Fubi-PA | | Descriptor: | 40S ribosomal protein S30, Ubiquitin carboxyl-terminal hydrolase 36, ZINC ION, ... | | Authors: | O'Dea, R, Gersch, M. | | Deposit date: | 2022-11-24 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular basis for ubiquitin/Fubi cross-reactivity in USP16 and USP36.
Nat.Chem.Biol., 19, 2023
|
|
4R17
 
 | | Ligand-induced aziridine-formation at subunit beta5 of the yeast 20S proteasome | | Descriptor: | (2S,3S)-3-methylaziridine-2-carboxylic acid, MAGNESIUM ION, Proteasome subunit alpha type-1, ... | | Authors: | Dubiella, C, Cui, H, Gersch, M, Brouwer, A.J, Sieber, S.A, Krueger, A, Liskamp, R, Groll, M. | | Deposit date: | 2014-08-04 | | Release date: | 2014-10-15 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Selective inhibition of the immunoproteasome by ligand-induced crosslinking of the active site.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4R18
 
 | | Ligand-induced Lys33-Thr1 crosslinking at subunit beta5 of the yeast 20S proteasome | | Descriptor: | ALPHA-AMINOBUTYRIC ACID, MAGNESIUM ION, PROTEASOME SUBUNIT ALPHA TYPE-1, ... | | Authors: | Dubiella, C, Cui, H, Gersch, M, Brouwer, A.J, Sieber, S.A, Krueger, A, Liskamp, R, Groll, M. | | Deposit date: | 2014-08-04 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Selective inhibition of the immunoproteasome by ligand-induced crosslinking of the active site.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
3V5I
 
 | |
