5YE5
 
 | | structure of endo-lysosomal TRPML1 channel inserting into nanodisc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, mammalian endo-lysosomal TRPML1 channel | | Authors: | Yang, M, Gao, N. | | Deposit date: | 2017-09-15 | | Release date: | 2017-11-15 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Cryo-EM structures of the mammalian endo-lysosomal TRPML1 channel elucidate the combined regulation mechanism
Protein Cell, 8, 2017
|
|
5YDZ
 
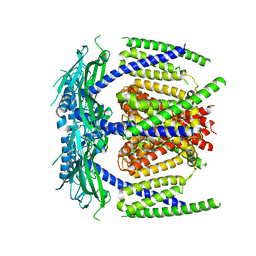 | |
8HFQ
 
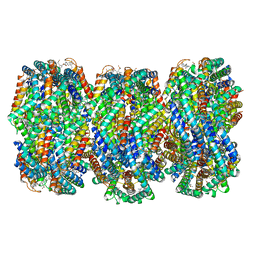 | | Cryo-EM structure of CpcL-PBS from cyanobacterium Synechocystis sp. PCC 6803 | | Descriptor: | C-phycocyanin alpha subunit, C-phycocyanin beta subunit, Ferredoxin--NADP reductase, ... | | Authors: | Zheng, L, Zhang, Z, Wang, H, Zheng, Z, Gao, N, Zhao, J. | | Deposit date: | 2022-11-11 | | Release date: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Cryo-EM and femtosecond spectroscopic studies provide mechanistic insight into the energy transfer in CpcL-phycobilisomes.
Nat Commun, 14, 2023
|
|
8INK
 
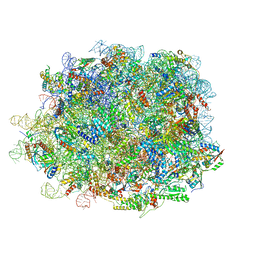 | | human nuclear pre-60S ribosomal particle - State D | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Zhang, Y, Gao, N. | | Deposit date: | 2023-03-10 | | Release date: | 2023-08-09 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Visualizing the nucleoplasmic maturation of human pre-60S ribosomal particles.
Cell Res., 33, 2023
|
|
8IE3
 
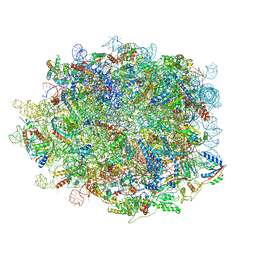 | | human nuclear pre-60S ribosomal particle - State E | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Zhang, Y, Gao, N. | | Deposit date: | 2023-02-15 | | Release date: | 2023-08-09 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Visualizing the nucleoplasmic maturation of human pre-60S ribosomal particles.
Cell Res., 33, 2023
|
|
8INF
 
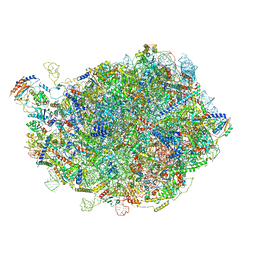 | |
8IDT
 
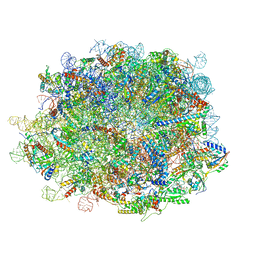 | | human nuclear pre-60S ribosomal particle - State G | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Zhang, Y, Gao, N. | | Deposit date: | 2023-02-14 | | Release date: | 2023-08-09 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Visualizing the nucleoplasmic maturation of human pre-60S ribosomal particles.
Cell Res., 33, 2023
|
|
8INE
 
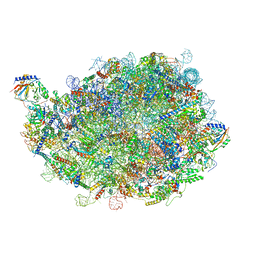 | |
8IDY
 
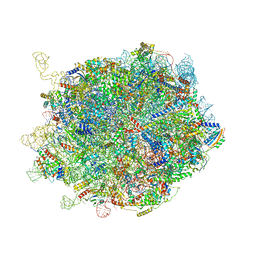 | |
8IPX
 
 | |
8IR3
 
 | |
8IPD
 
 | | human nuclear pre-60S ribosomal particle - State C | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Zhang, Y, Gao, N. | | Deposit date: | 2023-03-14 | | Release date: | 2023-08-09 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Visualizing the nucleoplasmic maturation of human pre-60S ribosomal particles.
Cell Res., 33, 2023
|
|
8IR1
 
 | | human nuclear pre-60S ribosomal particle - State A | | Descriptor: | 28S rRNA, 5.8S rRMA, 5S RNA, ... | | Authors: | Zhang, Y, Gao, N. | | Deposit date: | 2023-03-17 | | Release date: | 2023-08-09 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Visualizing the nucleoplasmic maturation of human pre-60S ribosomal particles.
Cell Res., 33, 2023
|
|
8IPY
 
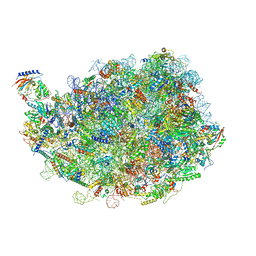 | |
5YE2
 
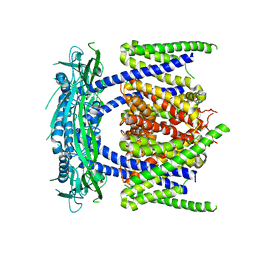 | |
8W8R
 
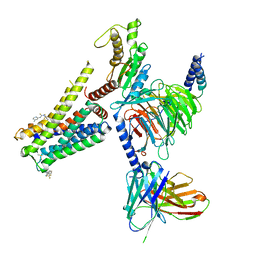 | | Cryo-EM structure of the AA-14-bound GPR101-Gs complex | | Descriptor: | 1-(4-methylpyridin-2-yl)-3-[3-(trifluoromethyl)phenyl]thiourea, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Sun, J.P, Yu, X, Gao, N, Yang, F, Wang, J.Y, Yang, Z, Guan, Y, Wang, G.P. | | Deposit date: | 2023-09-04 | | Release date: | 2024-01-03 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of GPR101-Gs enables identification of ligands with rejuvenating potential.
Nat.Chem.Biol., 20, 2024
|
|
8W8S
 
 | | Cryo-EM structure of the AA14-bound GPR101 complex | | Descriptor: | 1-(4-methylpyridin-2-yl)-3-[3-(trifluoromethyl)phenyl]thiourea, Probable G-protein coupled receptor 101 | | Authors: | Sun, J.P, Yu, X, Gao, N, Yang, F, Wang, J.Y, Yang, Z, Guan, Y, Wang, G.P. | | Deposit date: | 2023-09-04 | | Release date: | 2024-01-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of GPR101-Gs enables identification of ligands with rejuvenating potential.
Nat.Chem.Biol., 20, 2024
|
|
8W8Q
 
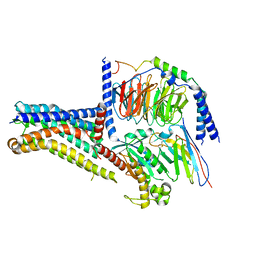 | | Cryo-EM structure of the GPR101-Gs complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Sun, J.P, Gao, N, Yu, X, Wang, G.P, Yang, F, Wang, J.Y, Yang, Z, Guan, Y. | | Deposit date: | 2023-09-04 | | Release date: | 2024-01-03 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structure of GPR101-Gs enables identification of ligands with rejuvenating potential.
Nat.Chem.Biol., 20, 2024
|
|
4HQM
 
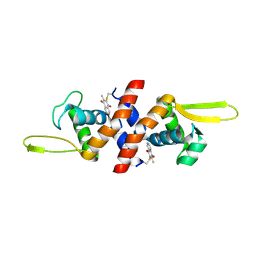 | | The crystal structure of QsrR-menadione complex | | Descriptor: | 2-methylnaphthalene-1,4-diol, QsrR protein | | Authors: | Ji, Q, Zhang, L, Jones, M.B, Sun, F, Deng, X, Liang, H, Brugarolas, P, Gao, N, Peterson, S.N, Lan, L, Bae, T, He, C. | | Deposit date: | 2012-10-25 | | Release date: | 2013-03-06 | | Last modified: | 2013-05-22 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Molecular mechanism of quinone signaling mediated through S-quinonization of a YodB family repressor QsrR.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6IRO
 
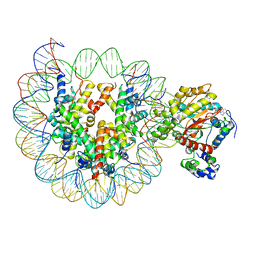 | | the crosslinked complex of ISWI-nucleosome in the ADP-bound state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (167-MER), Histone H2A, ... | | Authors: | Yan, L.J, Wu, H, Li, X.M, Gao, N, Chen, Z.C. | | Deposit date: | 2018-11-13 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of the ISWI-nucleosome complex reveal a conserved mechanism of chromatin remodeling.
Nat. Struct. Mol. Biol., 26, 2019
|
|
6ITC
 
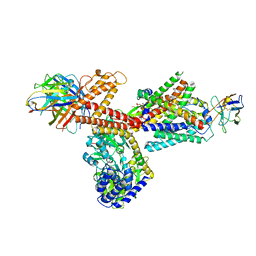 | | Structure of a substrate engaged SecA-SecY protein translocation machine | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Ma, C.Y, Wu, X.F, Sun, D.J, Park, E.Y, Rapoport, T.A, Gao, N, Long, L. | | Deposit date: | 2018-11-21 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structure of the substrate-engaged SecA-SecY protein translocation machine.
Nat Commun, 10, 2019
|
|
6K1P
 
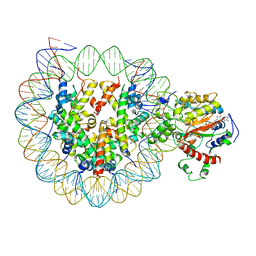 | | The complex of ISWI-nucleosome in the ADP.BeF-bound state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA (167-MER), ... | | Authors: | Yan, L.J, Wu, H, Li, X.M, Gao, N, Chen, Z.C. | | Deposit date: | 2019-05-10 | | Release date: | 2019-05-29 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Structures of the ISWI-nucleosome complex reveal a conserved mechanism of chromatin remodeling.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6JYL
 
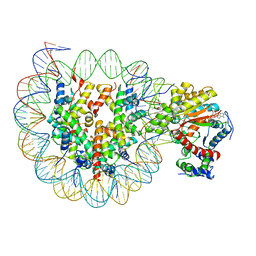 | | The crosslinked complex of ISWI-nucleosome in the ADP.BeF-bound state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA (167-MER), ... | | Authors: | Yan, L.J, Wu, H, Li, X.M, Gao, N, Chen, Z.C. | | Deposit date: | 2019-04-26 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Structures of the ISWI-nucleosome complex reveal a conserved mechanism of chromatin remodeling.
Nat.Struct.Mol.Biol., 26, 2019
|
|
5XY3
 
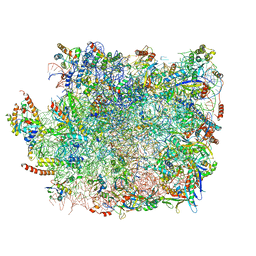 | | Large subunit of Trichomonas vaginalis ribosome | | Descriptor: | 25S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Li, Z, Guo, Q, Zheng, L, Ji, Y, Xie, Y, Lai, D, Lun, Z, Suo, X, Gao, N. | | Deposit date: | 2017-07-06 | | Release date: | 2017-08-30 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of the 80S ribosomes from human parasites Trichomonas vaginalis and Toxoplasma gondii
Cell Res., 27, 2017
|
|
5XYU
 
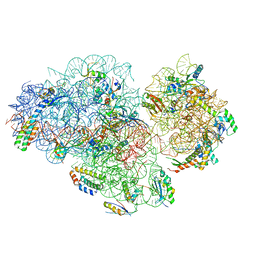 | | Small subunit of Mycobacterium smegmatis ribosome | | Descriptor: | 16S RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Li, Z, Zhang, Y, Zheng, L, Ge, X, Sanyal, S, Gao, N. | | Deposit date: | 2017-07-10 | | Release date: | 2017-09-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Cryo-EM structure of Mycobacterium smegmatis ribosome reveals two unidentified ribosomal proteins close to the functional centers.
Protein Cell, 9, 2018
|
|
