8FBZ
 
 | | Crystal Structure of apo human Glutathione Synthetase Y270E | | Descriptor: | GLYCEROL, Glutathione synthetase, SULFATE ION | | Authors: | Stanford, S.M, Santelli, E, Sankaran, B, Murali, R, Bottini, N. | | Deposit date: | 2022-11-30 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Targeting prostate tumor low-molecular weight tyrosine phosphatase for oxidation-sensitizing therapy.
Sci Adv, 10, 2024
|
|
8THM
 
 | |
5JNU
 
 | | Crystal structure of mouse Low-Molecular Weight Protein Tyrosine Phosphatase type A (LMPTP-A) complexed with phosphate | | Descriptor: | Low molecular weight phosphotyrosine protein phosphatase, PHOSPHATE ION | | Authors: | Stanford, S.M, Aleshin, A.E, Liddington, R.C, Bankston, L, Cadwell, G, Bottini, N. | | Deposit date: | 2016-04-30 | | Release date: | 2017-03-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.535 Å) | | Cite: | Diabetes reversal by inhibition of the low-molecular-weight tyrosine phosphatase.
Nat. Chem. Biol., 13, 2017
|
|
5JNS
 
 | | Crystal structure of human low molecular weight protein tyrosine phosphatase (LMPTP) type A complexed with phosphate | | Descriptor: | DIMETHYL SULFOXIDE, Low molecular weight phosphotyrosine protein phosphatase, PHOSPHATE ION | | Authors: | Stanford, S.M, Aleshin, A.E, Liddington, R.C, Bankston, L, Cadwell, G, Bottini, N. | | Deposit date: | 2016-04-30 | | Release date: | 2017-03-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Diabetes reversal by inhibition of the low-molecular-weight tyrosine phosphatase.
Nat. Chem. Biol., 13, 2017
|
|
5JNW
 
 | | Crystal structure of bovine low molecular weight protein tyrosine phosphatase (LMPTP) mutant (W49Y N50E) complexed with vanadate and uncompetitive inhibitor | | Descriptor: | 2-(4-{[3-(piperidin-1-yl)propyl]amino}quinolin-2-yl)benzonitrile, Low molecular weight phosphotyrosine protein phosphatase, VANADATE ION | | Authors: | Stanford, S.M, Aleshin, A.E, Liddington, R.C, Bankston, L, Cadwell, G, Bottini, N. | | Deposit date: | 2016-04-30 | | Release date: | 2017-03-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Diabetes reversal by inhibition of the low-molecular-weight tyrosine phosphatase.
Nat. Chem. Biol., 13, 2017
|
|
5JNV
 
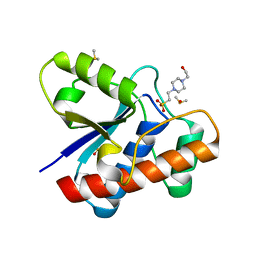 | | Crystal structure of bovine low molecular weight protein tyrosine phosphatase (LMPTP) mutant (W49Y N50E) complexed with HEPES | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DIMETHYL SULFOXIDE, Low molecular weight phosphotyrosine protein phosphatase | | Authors: | Stanford, S.M, Aleshin, A.E, Liddington, R.C, Bankston, L, Cadwell, G, Bottini, N. | | Deposit date: | 2016-04-30 | | Release date: | 2017-03-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Diabetes reversal by inhibition of the low-molecular-weight tyrosine phosphatase.
Nat. Chem. Biol., 13, 2017
|
|
5JNT
 
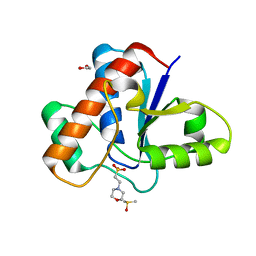 | | Crystal structure of human low molecular weight protein tyrosine phosphatase (LMPTP) type A complexed with MES | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DIMETHYL SULFOXIDE, ... | | Authors: | Stanford, S.M, Aleshin, A.E, Liddington, R.C, Bankston, L, Cadwell, G, Bottini, N. | | Deposit date: | 2016-04-30 | | Release date: | 2017-03-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Diabetes reversal by inhibition of the low-molecular-weight tyrosine phosphatase.
Nat. Chem. Biol., 13, 2017
|
|
5JNR
 
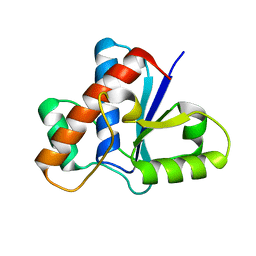 | | Crystal structure of human low molecular weight protein tyrosine phosphatase (LMPTP) type A | | Descriptor: | Low molecular weight phosphotyrosine protein phosphatase | | Authors: | Stanford, S.M, Aleshin, A.E, Liddington, R.C, Bankston, L, Cadwell, G, Bottini, N. | | Deposit date: | 2016-04-30 | | Release date: | 2017-03-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Diabetes reversal by inhibition of the low-molecular-weight tyrosine phosphatase.
Nat. Chem. Biol., 13, 2017
|
|
7KH8
 
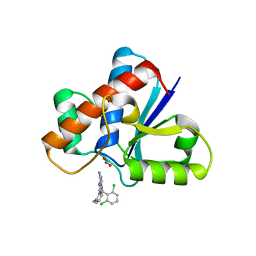 | | Human LMPTP in complex with inhibitor | | Descriptor: | 3-[(2,6-dichlorophenyl)methyl]-8-(2-methylphenyl)-3H-purin-6-amine, Low molecular weight phosphotyrosine protein phosphatase, NITRATE ION | | Authors: | Stanford, S.M, Diaz, M.A, Ardecky, R.J, Zou, J, Roosild, T, Holmes, Z.J, Hedrick, M.P, Rodiles, S, Santelli, E, Chung, T.D.Y, Jackson, M.R, Bottini, N, Pinkerton, A.B. | | Deposit date: | 2020-10-20 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Discovery of Orally Bioavailable Purine-Based Inhibitors of the Low-Molecular-Weight Protein Tyrosine Phosphatase.
J.Med.Chem., 64, 2021
|
|
3HHA
 
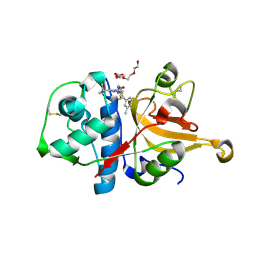 | | Crystal structure of cathepsin L in complex with AZ12878478 | | Descriptor: | ACETATE ION, Cathepsin L1, GLYCEROL, ... | | Authors: | Asaad, N, Bethel, P.A, Coulson, M.D, Dawson, J, Ford, S.J, Gerhardt, S, Grist, M, Hamlin, G.A, James, M.J, Jones, E.V, Karoutchi, G.I, Kenny, P.W, Morley, A.D, Oldham, K, Rankine, N, Ryan, D, Wells, S.L, Wood, L, Augustin, M, Krapp, S, Simader, H, Steinbacher, S. | | Deposit date: | 2009-05-15 | | Release date: | 2009-06-23 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Dipeptidyl nitrile inhibitors of Cathepsin L.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
1GH7
 
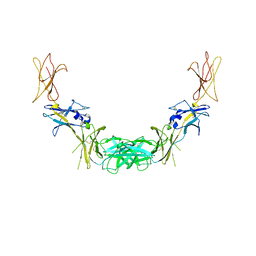 | | CRYSTAL STRUCTURE OF THE COMPLETE EXTRACELLULAR DOMAIN OF THE BETA-COMMON RECEPTOR OF IL-3, IL-5, AND GM-CSF | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CYTOKINE RECEPTOR COMMON BETA CHAIN, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Carr, P.D, Gustin, S.E, Church, A.P, Murphy, J.M, Ford, S.C, Mann, D.A, Woltring, D.M, Walker, I, Ollis, D.L, Young, I.G. | | Deposit date: | 2000-11-27 | | Release date: | 2001-11-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the complete extracellular domain of the common beta subunit of the human GM-CSF, IL-3, and IL-5 receptors reveals a novel dimer configuration.
Cell(Cambridge,Mass.), 104, 2001
|
|
2GYS
 
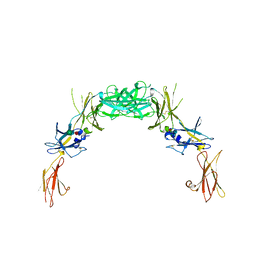 | | 2.7 A structure of the extracellular domains of the human beta common receptor involved in IL-3, IL-5, and GM-CSF signalling | | Descriptor: | Cytokine receptor common beta chain, alpha-L-fucopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Carr, P.D, Conlan, F, Ford, S, Ollis, D.L, Young, I.G. | | Deposit date: | 2006-05-09 | | Release date: | 2006-06-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An improved resolution structure of the human beta common receptor involved in IL-3, IL-5 and GM-CSF signalling which gives better definition of the high-affinity binding epitope.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
8BFB
 
 | | Sarkosyl-extracted AppNL-G-F Abeta42 fibril structure (Methoxy-X04-labelled mice) | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Wilkinson, M, Leistner, C, Burgess, A, Goodfellow, S, Deuchars, S, Ranson, N.A, Radford, S.E, Frank, R.A.W. | | Deposit date: | 2022-10-24 | | Release date: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The in-tissue molecular architecture of beta-amyloid pathology in the mammalian brain.
Nat Commun, 14, 2023
|
|
8BFA
 
 | | Sarkosyl-extracted AppNL-G-F Abeta42 fibril structure | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Wilkinson, M, Leistner, C, Burgess, A, Goodfellow, S, Deuchars, S, Ranson, N.A, Radford, S.E, Frank, R.A.W. | | Deposit date: | 2022-10-24 | | Release date: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The in-tissue molecular architecture of beta-amyloid pathology in the mammalian brain.
Nat Commun, 14, 2023
|
|
7BM5
 
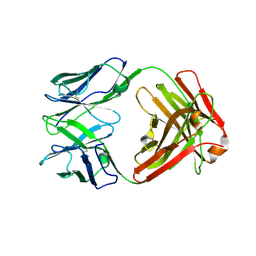 | | Crystal structure of Fab1, the Fab fragment of the anti-BamA monoclonal antibody MAB1 | | Descriptor: | Fab1 heavy chain, Fab1 light chain | | Authors: | White, P, Storek, K.M, Rutherford, S.T, Radford, S.E. | | Deposit date: | 2021-01-19 | | Release date: | 2021-06-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The role of membrane destabilisation and protein dynamics in BAM catalysed OMP folding.
Nat Commun, 12, 2021
|
|
8BVQ
 
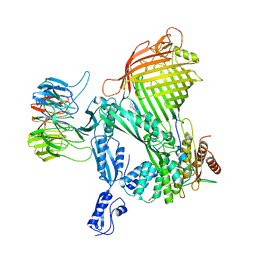 | | E. coli BAM complex (BamABCDE) bound to darobactin B | | Descriptor: | Darobactin-B, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Horne, J.E, Fenn, K.L, Radford, S.E, Ranson, N.A. | | Deposit date: | 2022-12-05 | | Release date: | 2023-05-24 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Darobactin B Stabilises a Lateral-Closed Conformation of the BAM Complex in E. coli Cells.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8BWC
 
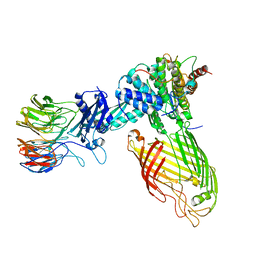 | | E. coli BAM complex (BamABCDE) wild-type | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Machin, J.M, Radford, S.E, Ranson, N.A. | | Deposit date: | 2022-12-06 | | Release date: | 2023-05-24 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Darobactin B Stabilises a Lateral-Closed Conformation of the BAM Complex in E. coli Cells.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8OH2
 
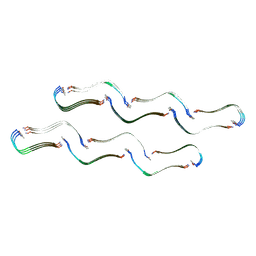 | | Structure of the Tau-PAM4 Type 1 amyloid fibril | | Descriptor: | Microtubule-associated protein tau | | Authors: | Wilkinson, M, Louros, N, Tsaka, G, Ramakers, M, Morelli, C, Garcia, T, Gallardo, R.U, D'Haeyer, S, Goossens, V, Audenaert, D, Thal, D.R, Ranson, N.A, Radford, S.E, Rousseau, F, Schymkowitz, J. | | Deposit date: | 2023-03-20 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Local structural preferences in shaping tau amyloid polymorphism.
Nat Commun, 15, 2024
|
|
8OHP
 
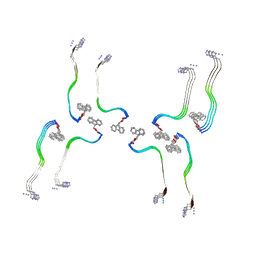 | | Structure of the Fmoc-Tau-PAM4 Type 3 amyloid fibril | | Descriptor: | Microtubule-associated protein tau | | Authors: | Wilkinson, M, Louros, N, Tsaka, G, Ramakers, M, Morelli, C, Garcia, T, Gallardo, R.U, D'Haeyer, S, Goossens, V, Audenaert, D, Thal, D.R, Ranson, N.A, Radford, S.E, Rousseau, F, Schymkowitz, J. | | Deposit date: | 2023-03-21 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Local structural preferences in shaping tau amyloid polymorphism.
Nat Commun, 15, 2024
|
|
8OI0
 
 | | Structure of the Fmoc-Tau-PAM4 Type 4 amyloid fibril | | Descriptor: | Microtubule-associated protein tau | | Authors: | Wilkinson, M, Louros, N, Tsaka, G, Ramakers, M, Morelli, C, Garcia, T, Gallardo, R.U, D'Haeyer, S, Goossens, V, Audenaert, D, Thal, D.R, Ranson, N.A, Radford, S.E, Rousseau, F, Schymkowitz, J. | | Deposit date: | 2023-03-21 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Local structural preferences in shaping tau amyloid polymorphism.
Nat Commun, 15, 2024
|
|
8OHI
 
 | | Structure of the Fmoc-Tau-PAM4 Type 2 amyloid fibril | | Descriptor: | Microtubule-associated protein tau | | Authors: | Wilkinson, M, Louros, N, Tsaka, G, Ramakers, M, Morelli, C, Garcia, T, Gallardo, R.U, D'Haeyer, S, Goossens, V, Audenaert, D, Thal, D.R, Ranson, N.A, Radford, S.E, Rousseau, F, Schymkowitz, J. | | Deposit date: | 2023-03-21 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Local structural preferences in shaping tau amyloid polymorphism.
Nat Commun, 15, 2024
|
|
6XOD
 
 | | Crystal structure of the PEX4-PEX22 protein complex from Arabidopsis thaliana | | Descriptor: | Peroxisome biogenesis protein 22, Protein PEROXIN-4 | | Authors: | Olmos Jr, J.L, Bradford, S.E, Miller, M.D, Xu, W, Wright, Z.J, Bartel, B, Phillips Jr, G.N. | | Deposit date: | 2020-07-06 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The Structure of the Arabidopsis PEX4-PEX22 Peroxin Complex-Insights Into Ubiquitination at the Peroxisomal Membrane
Front Cell Dev Biol, 10, 2022
|
|
5DLS
 
 | | Identification of Novel, in vivo Active Chk1 Inhibitors Utilizing Structure Guided Drug Design | | Descriptor: | 1-benzyl-N-(5-{5-[3-(dimethylamino)-2,2-dimethylpropoxy]-1H-indol-2-yl}-6-oxo-1,6-dihydropyridin-3-yl)-1H-pyrazole-4-carboxamide, SULFATE ION, Serine/threonine-protein kinase Chk1 | | Authors: | Massey, A.J, Stokes, S, Browne, H, Foloppe, N, Fiumana, A, Scrace, S, Fallowfield, M, Bedford, S, Webb, P, Baker, L.M, Christie, M, Drysdale, M.J, Wood, M. | | Deposit date: | 2015-09-07 | | Release date: | 2015-10-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Identification of novel, in vivo active Chk1 inhibitors utilizing structure guided drug design.
Oncotarget, 6, 2015
|
|
6S0L
 
 | | Structure of the A2A adenosine receptor determined at SwissFEL using native-SAD at 4.57 keV from all available diffraction patterns | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | Authors: | Nass, K, Cheng, R, Vera, L, Mozzanica, A, Redford, S, Ozerov, D, Basu, S, James, D, Knopp, G, Cirelli, C, Martiel, I, Casadei, C, Weinert, T, Nogly, P, Skopintsev, P, Usov, I, Leonarski, F, Geng, T, Rappas, M, Dore, A.S, Cooke, R, Nasrollahi Shirazi, S, Dworkowski, F, Sharpe, M, Olieric, N, Steinmetz, M.O, Schertler, G, Abela, R, Patthey, L, Schmitt, B, Hennig, M, Standfuss, J, Wang, M, Milne, J.C. | | Deposit date: | 2019-06-17 | | Release date: | 2020-07-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Advances in long-wavelength native phasing at X-ray free-electron lasers.
Iucrj, 7, 2020
|
|
6S19
 
 | | Structure of thaumatin determined at SwissFEL using native-SAD at 4.57 keV from all available diffraction patterns | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Nass, K, Cheng, R, Vera, L, Mozzanica, A, Redford, S, Ozerov, D, Basu, S, James, D, Knopp, G, Cirelli, C, Martiel, I, Casadei, C, Weinert, T, Nogly, P, Skopintsev, P, Usov, I, Leonarski, F, Geng, T, Rappas, M, Dore, A.S, Cooke, R, Nasrollahi Shirazi, S, Dworkowski, F, Sharpe, M, Olieric, N, Steinmetz, M.O, Schertler, G, Abela, R, Patthey, L, Schmitt, B, Hennig, M, Standfuss, J, Wang, M, Milne, J.Ch. | | Deposit date: | 2019-06-18 | | Release date: | 2020-07-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Advances in long-wavelength native phasing at X-ray free-electron lasers.
Iucrj, 7, 2020
|
|
