4YTA
 
 | | BOND LENGTH ANALYSIS OF ASP, GLU AND HIS RESIDUES IN TRYPSIN AT 1.2A RESOLUTION | | 分子名称: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | 著者 | Fisher, S.J, Helliwell, J.R, Blakeley, M.P, Cianci, M, McSweeny, S. | | 登録日 | 2015-03-17 | | 公開日 | 2015-05-27 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Protonation-state determination in proteins using high-resolution X-ray crystallography: effects of resolution and completeness.
Acta Crystallogr. D Biol. Crystallogr., 68, 2012
|
|
4N0X
 
 | |
4Q49
 
 | |
4NY6
 
 | | Neutron structure of leucine and valine methyl protonated type III antifreeze | | 分子名称: | Type-3 ice-structuring protein HPLC 12 | | 著者 | Fisher, S.J, Blakeley, M.P, Howard, E.I, Petite-Haertlein, I, Haertlein, M, Mitschler, A, Cousido-Siah, A, Salvaya, A.G, Popov, A, Muller-Dieckmann, C, Petrova, T, Podjarny, A.D. | | 登録日 | 2013-12-10 | | 公開日 | 2014-12-24 | | 最終更新日 | 2024-02-28 | | 実験手法 | NEUTRON DIFFRACTION (1.05 Å), X-RAY DIFFRACTION | | 主引用文献 | Perdeuteration: improved visualization of solvent structure in neutron macromolecular crystallography.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2NXS
 
 | |
2NWO
 
 | |
2NWY
 
 | |
4G0C
 
 | | Neutron structure of acetazolamide-bound human carbonic anhydrase II reveal molecular details of drug binding. | | 分子名称: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, Carbonic anhydrase 2, ZINC ION | | 著者 | Fisher, S.Z, McKenna, R, Aggarwal, M. | | 登録日 | 2012-07-09 | | 公開日 | 2012-09-19 | | 最終更新日 | 2023-09-13 | | 実験手法 | NEUTRON DIFFRACTION (2 Å) | | 主引用文献 | Neutron diffraction of acetazolamide-bound human carbonic anhydrase II reveals atomic details of drug binding.
J.Am.Chem.Soc., 134, 2012
|
|
8IGF
 
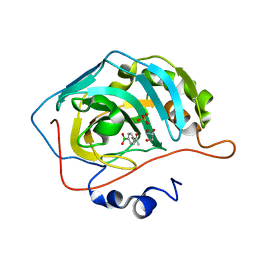 | | Crystal Structure of Human Carbonic Anhydrase II In-complex with 4-Acetylphenylboronic acid at 2.6 A Resolution | | 分子名称: | (4-ethanoylphenyl)boronic acid, Carbonic anhydrase 2, GLYCEROL, ... | | 著者 | Rasheed, S, Huda, N, Fisher, S.Z, Falke, S, Gul, S, Ahmad, M.S, Choudhary, M.I. | | 登録日 | 2023-02-20 | | 公開日 | 2024-02-28 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Identification, crystallization, and first X-ray structure analyses of phenyl boronic acid-based inhibitors of human carbonic anhydrase-II.
Int.J.Biol.Macromol., 267, 2024
|
|
7ORW
 
 | | Non-structural protein 10 (nsp10) from SARS CoV-2 in complex with fragment VT00265 | | 分子名称: | 1H-benzimidazol-4-amine, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | 著者 | Talibov, V.O, Kozielski, F, Sele, C, Lou, J, Dong, D, Wang, Q, Shi, X, Nyblom, M, Rogstam, A, Krojer, T, Knecht, W, Fisher, S.Z. | | 登録日 | 2021-06-06 | | 公開日 | 2021-10-20 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Identification of fragments binding to SARS-CoV-2 nsp10 reveals ligand-binding sites in conserved interfaces between nsp10 and nsp14/nsp16.
Rsc Chem Biol, 3, 2022
|
|
7ORU
 
 | | Non-structural protein 10 (nsp10) from SARS CoV-2 in complex with fragment VT00221 | | 分子名称: | CHLORIDE ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | 著者 | Talibov, V.O, Kozielski, F, Sele, C, Lou, J, Dong, D, Wang, Q, Shi, X, Nyblom, M, Rogstam, A, Krojer, T, Knecht, W, Fisher, S.Z. | | 登録日 | 2021-06-06 | | 公開日 | 2021-10-20 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Identification of fragments binding to SARS-CoV-2 nsp10 reveals ligand-binding sites in conserved interfaces between nsp10 and nsp14/nsp16.
Rsc Chem Biol, 3, 2022
|
|
7ORV
 
 | | Non-structural protein 10 (nsp10) from SARS CoV-2 in complex with fragment VT00239 | | 分子名称: | CHLORIDE ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | 著者 | Talibov, V.O, Kozielski, F, Sele, C, Lou, J, Dong, D, Wang, Q, Shi, X, Nyblom, M, Rogstam, A, Krojer, T, Knecht, W, Fisher, S.Z. | | 登録日 | 2021-06-06 | | 公開日 | 2021-10-20 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Identification of fragments binding to SARS-CoV-2 nsp10 reveals ligand-binding sites in conserved interfaces between nsp10 and nsp14/nsp16.
Rsc Chem Biol, 3, 2022
|
|
7ORR
 
 | | Non-structural protein 10 (nsp10) from SARS CoV-2 in complex with fragment VT00022 | | 分子名称: | 4-PHENYL-1H-IMIDAZOLE, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | 著者 | Talibov, V.O, Kozielski, F, Sele, C, Lou, J, Dong, D, Wang, Q, Shi, X, Nyblom, M, Rogstam, A, Krojer, T, Knecht, W, Fisher, S.Z. | | 登録日 | 2021-06-06 | | 公開日 | 2021-10-20 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Identification of fragments binding to SARS-CoV-2 nsp10 reveals ligand-binding sites in conserved interfaces between nsp10 and nsp14/nsp16.
Rsc Chem Biol, 3, 2022
|
|
6YMA
 
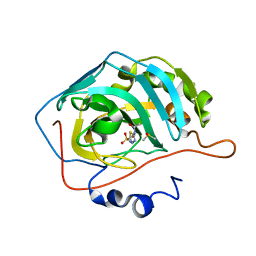 | | MicroED structure of acetazolamide-bound human carbonic anhydrase II | | 分子名称: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, DIMETHYL SULFOXIDE, ZINC ION, ... | | 著者 | Clabbers, M.T.B, Fisher, S.Z, Coincon, M, Zou, X, Xu, H. | | 登録日 | 2020-04-08 | | 公開日 | 2020-08-12 | | 最終更新日 | 2024-01-24 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (2.5 Å) | | 主引用文献 | Visualizing drug binding interactions using microcrystal electron diffraction.
Commun Biol, 3, 2020
|
|
8JEE
 
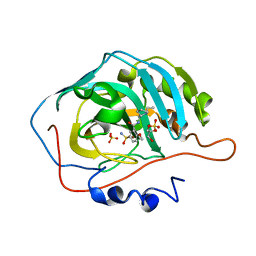 | | Crystal Structure of Human Carbonic Anhydrase II In-complex with Levosulpiride at 2.96 A Resolution | | 分子名称: | Carbonic anhydrase 2, GLYCEROL, Levosulpiride, ... | | 著者 | Rasheed, S, Huda, N, Falke, S, Fisher, S.Z, Ahmad, M.S. | | 登録日 | 2023-05-15 | | 公開日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (2.96 Å) | | 主引用文献 | Crystal Structure of Human Carbonic Anhydrase II In-complex with Levosulpiride at 2.96 A Resolution
To Be Published
|
|
6YMB
 
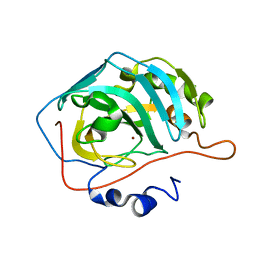 | | MicroED structure of human carbonic anhydrase II | | 分子名称: | ZINC ION, carbonic anhydrase 2 | | 著者 | Clabbers, M.T.B, Fisher, S.Z, Coincon, M, Zou, X, Xu, H. | | 登録日 | 2020-04-08 | | 公開日 | 2020-08-12 | | 最終更新日 | 2024-01-24 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (2.5 Å) | | 主引用文献 | Visualizing drug binding interactions using microcrystal electron diffraction.
Commun Biol, 3, 2020
|
|
6ZCT
 
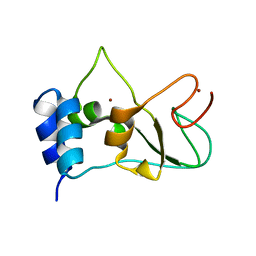 | | Nonstructural protein 10 (nsp10) from SARS CoV-2 | | 分子名称: | ZINC ION, nsp10 | | 著者 | Rogstam, A, Nyblom, M, Christensen, S, Sele, C, Lindvall, T, Rasmussen, A.A, Andre, I, Fisher, S.Z, Knecht, W, Kozielski, F. | | 登録日 | 2020-06-12 | | 公開日 | 2020-06-24 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Crystal Structure of Non-Structural Protein 10 from Severe Acute Respiratory Syndrome Coronavirus-2.
Int J Mol Sci, 21, 2020
|
|
4LNI
 
 | | B. subtilis glutamine synthetase structures reveal large active site conformational changes and basis for isoenzyme specific regulation: structure of the transition state complex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Glutamine synthetase, L-METHIONINE-S-SULFOXIMINE PHOSPHATE, ... | | 著者 | Schumacher, M.A, Chinnam, N, Tonthat, N, Fisher, S, Wray, L. | | 登録日 | 2013-07-11 | | 公開日 | 2013-11-06 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.5793 Å) | | 主引用文献 | Structures of the Bacillus subtilis Glutamine Synthetase Dodecamer Reveal Large Intersubunit Catalytic Conformational Changes Linked to a Unique Feedback Inhibition Mechanism.
J.Biol.Chem., 288, 2013
|
|
5ICR
 
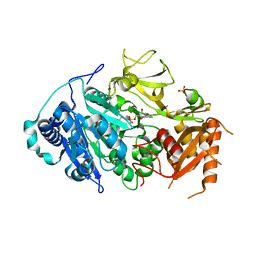 | | 2.25 Angstrom Resolution Crystal Structure of Fatty-Acid-CoA Ligase (FadD32) from Mycobacterium smegmatis in complex with Inhibitor 5'-O-[(11-phenoxyundecanoyl)sulfamoyl]adenosine. | | 分子名称: | 5'-O-[(11-phenoxyundecanoyl)sulfamoyl]adenosine, Acyl-CoA synthase, CHLORIDE ION, ... | | 著者 | Minasov, G, Shuvalova, L, Hung, D, Fisher, S.L, Edelstein, J, Kiryukhina, O, Dubrovska, I, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2016-02-23 | | 公開日 | 2016-04-06 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | 2.25 Angstrom Resolution Crystal Structure of Fatty-Acid-CoA Ligase (FadD32) from Mycobacterium smegmatis in complex with Inhibitor 5'-O-[(11-phenoxyundecanoyl)sulfamoyl]adenosine.
To Be Published
|
|
4LNK
 
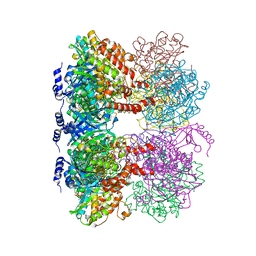 | | B. subtilis glutamine synthetase structures reveal large active site conformational changes and basis for isoenzyme specific regulation: structure of GS-glutamate-AMPPCP complex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, GLUTAMIC ACID, Glutamine synthetase, ... | | 著者 | Schumacher, M.A, Chinnam, N, Tonthat, N, Fisher, S, Wray, L. | | 登録日 | 2013-07-11 | | 公開日 | 2013-10-30 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.87 Å) | | 主引用文献 | Structures of the Bacillus subtilis Glutamine Synthetase Dodecamer Reveal Large Intersubunit Catalytic Conformational Changes Linked to a Unique Feedback Inhibition Mechanism.
J.Biol.Chem., 288, 2013
|
|
4LNF
 
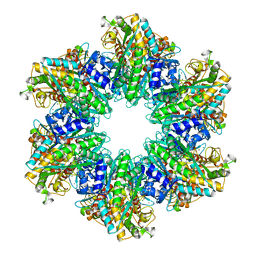 | | B. subtilis glutamine synthetase structures reveal large active site conformational changes and basis for isoenzyme specific regulation: structure of GS-Q | | 分子名称: | GLUTAMINE, Glutamine synthetase, MAGNESIUM ION, ... | | 著者 | Schumacher, M.A, Chinnam, N, Tonthat, N, Fisher, S, Wray, L. | | 登録日 | 2013-07-11 | | 公開日 | 2013-11-13 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.949 Å) | | 主引用文献 | Structures of the Bacillus subtilis Glutamine Synthetase Dodecamer Reveal Large Intersubunit Catalytic Conformational Changes Linked to a Unique Feedback Inhibition Mechanism.
J.Biol.Chem., 288, 2013
|
|
4LNN
 
 | | B. subtilis glutamine synthetase structures reveal large active site conformational changes and basis for isoenzyme specific regulation: structure of apo form of GS | | 分子名称: | Glutamine synthetase, MAGNESIUM ION, SULFATE ION | | 著者 | Schumacher, M.A, Chinnam, N, Tonthat, N, Fisher, S, Wray, L. | | 登録日 | 2013-07-11 | | 公開日 | 2013-11-13 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structures of the Bacillus subtilis Glutamine Synthetase Dodecamer Reveal Large Intersubunit Catalytic Conformational Changes Linked to a Unique Feedback Inhibition Mechanism.
J.Biol.Chem., 288, 2013
|
|
4LNO
 
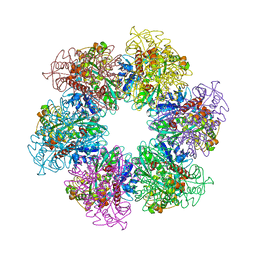 | | B. subtilis glutamine synthetase structures reveal large active site conformational changes and basis for isoenzyme specific regulation: form two of GS-1 | | 分子名称: | GLUTAMINE, Glutamine synthetase, MAGNESIUM ION | | 著者 | Schumacher, M.A, Chinnam, N, Tonthat, N, Fisher, S, Wray, L. | | 登録日 | 2013-07-11 | | 公開日 | 2013-11-13 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structures of the Bacillus subtilis Glutamine Synthetase Dodecamer Reveal Large Intersubunit Catalytic Conformational Changes Linked to a Unique Feedback Inhibition Mechanism.
J.Biol.Chem., 288, 2013
|
|
3V3F
 
 | |
3V3J
 
 | |
