7ZJ3
 
 | | Structure of TRIM2 RING domain in complex with UBE2D1~Ub conjugate | | Descriptor: | Polyubiquitin-C, Tripartite motif-containing protein 2, Ubiquitin-conjugating enzyme E2 D1, ... | | Authors: | Esposito, D, Garza-Garcia, A, Dudley-Fraser, J, Rittinger, K. | | Deposit date: | 2022-04-08 | | Release date: | 2022-11-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Divergent self-association properties of paralogous proteins TRIM2 and TRIM3 regulate their E3 ligase activity.
Nat Commun, 13, 2022
|
|
1LR1
 
 | | Solution Structure of the Oligomerization Domain of the Bacterial Chromatin-Structuring Protein H-NS | | Descriptor: | dna-binding protein h-ns | | Authors: | Esposito, D, Petrovic, A, Harris, R, Ono, S, Eccleston, J, Mbabaali, A, Haq, I, Higgins, C.F, Hinton, J.C.D, Driscoll, P.C, Ladbury, J.E. | | Deposit date: | 2002-05-14 | | Release date: | 2003-01-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | H-NS Oligomerization Domain Structure Reveals the Mechanism for High Order
Self-association of the Intact Protein
J.Mol.Biol., 324, 2002
|
|
6EYR
 
 | |
6EYT
 
 | |
8CR0
 
 | | Crystal structure of human carbonic anhydrase II in complex with a triazolyl benzoxaborole inhibitor | | Descriptor: | 1-[1,1-bis(oxidanyl)-3~{H}-2,1$l^{4}-benzoxaborol-6-yl]-4-[(4-fluoranylphenoxy)methyl]-1,2,3-triazole, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Alterio, V, De Simone, G, Esposito, D. | | Deposit date: | 2023-03-07 | | Release date: | 2023-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | 6-Substituted Triazolyl Benzoxaboroles as Selective Carbonic Anhydrase Inhibitors: In Silico Design, Synthesis, and X-ray Crystallography.
J.Med.Chem., 66, 2023
|
|
6I9H
 
 | |
6EQU
 
 | | X-Ray crystal structure of the human carbonic anhydrase II adduct with a membrane-impermeant inhibitor | | Descriptor: | 4-[2-(2,4,6-triphenylpyridin-1-ium-1-yl)ethyl]benzenesulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Alterio, V, De Simone, G, Esposito, D. | | Deposit date: | 2017-10-15 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of the human carbonic anhydrase II adduct with 1-(4-sulfamoylphenyl-ethyl)-2,4,6-triphenylpyridinium perchlorate, a membrane-impermeant, isoform selective inhibitor.
J Enzyme Inhib Med Chem, 33, 2018
|
|
5O07
 
 | |
5FEY
 
 | | TRIM32 RING | | Descriptor: | E3 ubiquitin-protein ligase TRIM32, ZINC ION | | Authors: | Rittinger, K, Esposito, D, Koliopoulos, M.G. | | Deposit date: | 2015-12-17 | | Release date: | 2016-05-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Functional role of TRIM E3 ligase oligomerization and regulation of catalytic activity.
Embo J., 35, 2016
|
|
5JQT
 
 | | Crystal structure of human carbonic anhydrase II in complex with Benzoxaborole at pH 7.4 | | Descriptor: | 1,1-dihydroxy-1,3-dihydro-2,1-benzoxaborol-1-ium, 2,1-benzoxaborol-1(3H)-ol, 4-(HYDROXYMERCURY)BENZOIC ACID, ... | | Authors: | Alterio, V, Esposito, D, Di Fiore, A, De Simone, G. | | Deposit date: | 2016-05-05 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Benzoxaborole as a new chemotype for carbonic anhydrase inhibition.
Chem.Commun.(Camb.), 52, 2016
|
|
5JQ0
 
 | | Crystal structure of human carbonic anhydrase II in complex with Benzoxaborole at pH=8.7 | | Descriptor: | 1,1-dihydroxy-1,3-dihydro-2,1-benzoxaborol-1-ium, Carbonic anhydrase 2, ZINC ION | | Authors: | Alterio, V, Esposito, D, Di Fiore, A, De Simone, G. | | Deposit date: | 2016-05-04 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Benzoxaborole as a new chemotype for carbonic anhydrase inhibition.
Chem.Commun.(Camb.), 52, 2016
|
|
5LMD
 
 | | The crystal structure of hCA II in complex with a benzoxaborole inhibitor | | Descriptor: | 1-[7,7-bis(oxidanyl)-8-oxa-7-boranuidabicyclo[4.3.0]nona-1,3,5-trien-4-yl]-3-(2-methoxy-5-methyl-phenyl)urea, Carbonic anhydrase 2, ZINC ION | | Authors: | De Simone, G, Alterio, V, Esposito, D, Di Fiore, A. | | Deposit date: | 2016-07-29 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Benzoxaborole as a new chemotype for carbonic anhydrase inhibition.
Chem.Commun.(Camb.), 52, 2016
|
|
5FER
 
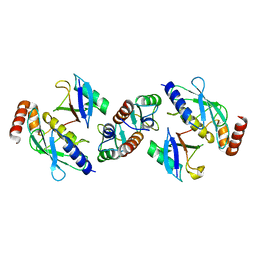 | | Complex of TRIM25 RING with UbcH5-Ub | | Descriptor: | E3 ubiquitin/ISG15 ligase TRIM25, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-conjugating enzyme E2 D1, ... | | Authors: | Rittinger, K, Koliopoulos, M.G, Esposito, D. | | Deposit date: | 2015-12-17 | | Release date: | 2016-05-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Functional role of TRIM E3 ligase oligomerization and regulation of catalytic activity.
Embo J., 35, 2016
|
|
8C0Q
 
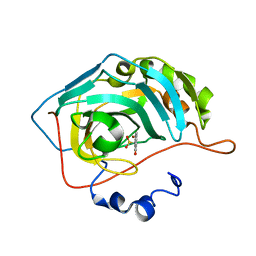 | | Crystal structure of human carbonic anhydrase II in complex with a coumarin derivative. | | Descriptor: | (4-methyl-2-oxidanylidene-chromen-7-yl)methanesulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Alterio, V, De Simone, G, Esposito, D. | | Deposit date: | 2022-12-19 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | A Combined in Silico and Structural Study Opens New Perspectives on Aliphatic Sulfonamides, a Still Poorly Investigated Class of CA Inhibitors.
Biology (Basel), 12, 2023
|
|
8C0R
 
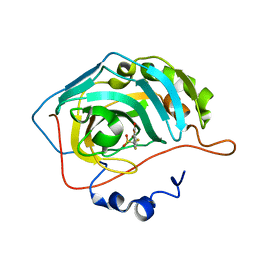 | | Crystal structure of human carbonic anhydrase II in complex with a coumarin derivative. | | Descriptor: | Carbonic anhydrase 2, ZINC ION, bis(fluoranyl)-(4-methyl-2-oxidanylidene-chromen-7-yl)methanesulfonamide | | Authors: | Alterio, V, De Simone, G, Esposito, D. | | Deposit date: | 2022-12-19 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | A Combined in Silico and Structural Study Opens New Perspectives on Aliphatic Sulfonamides, a Still Poorly Investigated Class of CA Inhibitors.
Biology (Basel), 12, 2023
|
|
6RPS
 
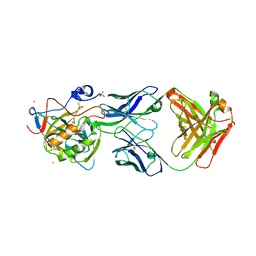 | | X-ray crystal structure of carbonic anhydrase XII complexed with a theranostic monoclonal antibody fragment | | Descriptor: | ACETATE ION, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Alterio, V, Esposito, D, De Simone, G. | | Deposit date: | 2019-05-14 | | Release date: | 2019-11-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Biochemical and Structural Insights into Carbonic Anhydrase XII/Fab6A10 Complex.
J.Mol.Biol., 431, 2019
|
|
6SDT
 
 | | HUMAN CARBONIC ANHYDRASE VII IN COMPLEX WITH A SULFONAMIDE INHIBITOR | | Descriptor: | Carbonic anhydrase 7, ZINC ION, phenyl-(4-sulfamoylphenoxy)phosphinic acid | | Authors: | Alterio, V, De Simone, G, Esposito, D. | | Deposit date: | 2019-07-29 | | Release date: | 2020-06-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Phenyl(thio)phosphon(amid)ate Benzenesulfonamides as Potent and Selective Inhibitors of Human Carbonic Anhydrases II and VII Counteract Allodynia in a Mouse Model of Oxaliplatin-Induced Neuropathy.
J.Med.Chem., 63, 2020
|
|
6SDS
 
 | | HUMAN CARBONIC ANHYDRASE II IN COMPLEX WITH A SULFONAMIDE INHIBITOR | | Descriptor: | Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ZINC ION, ... | | Authors: | Alterio, V, De Simone, G, Esposito, D. | | Deposit date: | 2019-07-29 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Phenyl(thio)phosphon(amid)ate Benzenesulfonamides as Potent and Selective Inhibitors of Human Carbonic Anhydrases II and VII Counteract Allodynia in a Mouse Model of Oxaliplatin-Induced Neuropathy.
J.Med.Chem., 63, 2020
|
|
2BYE
 
 | | NMR solution structure of phospholipase c epsilon RA 1 domain | | Descriptor: | PHOSPHOLIPASE C, EPSILON 1 | | Authors: | Bunney, T.D, Harris, R, Gandarillas, N.L, Josephs, M.B, Roe, S.M, Paterson, H.F, Rodrigues-Lima, F, Esposito, D, Gieschik, P, Pearl, L.H, Driscoll, P.C, Katan, M. | | Deposit date: | 2005-08-01 | | Release date: | 2006-02-22 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural and Mechanistic Insights Into Ras Association Domains of Phospholipase C Epsilon.
Mol.Cell, 21, 2006
|
|
2BYF
 
 | | NMR solution structure of phospholipase c epsilon RA 2 domain | | Descriptor: | PHOSPHOLIPASE C, EPSILON 1 | | Authors: | Bunney, T.D, Harris, R, Gandarillas, N.L, Josephs, M.B, Roe, S.M, Paterson, H.F, Rodrigues-Lima, F, Esposito, D, Gieschik, P, Pearl, L.H, Driscoll, P.C, Katan, M. | | Deposit date: | 2005-08-01 | | Release date: | 2006-02-22 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural and Mechanistic Insights Into Ras Association Domains of Phospholipase C Epsilon.
Mol.Cell, 21, 2006
|
|
1Q2Z
 
 | | The 3D solution structure of the C-terminal region of Ku86 | | Descriptor: | ATP-dependent DNA helicase II, 80 kDa subunit | | Authors: | Harris, R, Esposito, D, Sankar, A, Maman, J.D, Hinks, J.A, Pearl, L.H, Driscoll, P.C. | | Deposit date: | 2003-07-28 | | Release date: | 2004-01-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The 3D Solution Structure of the C-terminal Region of Ku86 (Ku86CTR)
J.Mol.Biol., 335, 2004
|
|
6GGO
 
 | | Crystal structure of Salmonella zinc metalloprotease effector GtgA | | Descriptor: | Bacteriophage virulence determinant, CHLORIDE ION, ZINC ION | | Authors: | Jennings, E, Esposito, D, Rittinger, K, Thurston, T. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-function analyses of the bacterial zinc metalloprotease effector protein GtgA uncover key residues required for deactivating NF-kappa B.
J. Biol. Chem., 293, 2018
|
|
6GGR
 
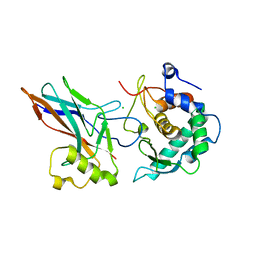 | | Crystal structure of Salmonella zinc metalloprotease effector GtgA in complex with p65 | | Descriptor: | Bacteriophage virulence determinant, CHLORIDE ION, Transcription factor p65 | | Authors: | Jennings, E, Esposito, D, Rittinger, K, Thurston, T. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Structure-function analyses of the bacterial zinc metalloprotease effector protein GtgA uncover key residues required for deactivating NF-kappa B.
J. Biol. Chem., 293, 2018
|
|
7TPR
 
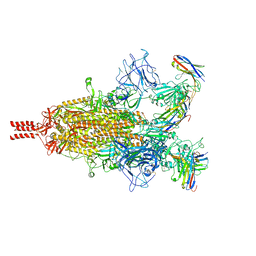 | | Camel nanobodies 7A3 and 8A2 broadly neutralize SARS-CoV-2 variants | | Descriptor: | Nanobody 7A3, Nanobody 8A2, Spike glycoprotein | | Authors: | Butay, K.J, Zhu, J, Dandey, V.P, Hong, J, Kwon, H.J, Chen, C.Z, Duan, Z, Li, D, Ren, H, Liang, T, Martin, N, Esposito, D, Ortega-Rodriguez, U, Xu, M, Xie, H, Ho, M, Cachau, R, Borgnia, M.J. | | Deposit date: | 2022-01-25 | | Release date: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (2.39 Å) | | Cite: | Camel nanobodies broadly neutralize SARS-CoV-2 variants
bioRxiv, 2021
|
|
2JP0
 
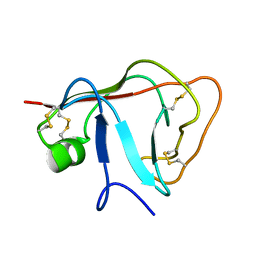 | | Solution structure of the N-terminal extraceullular domain of the lymphocyte receptor CD5 calculated using inferential structure determination (ISD) | | Descriptor: | T-cell surface glycoprotein CD5 | | Authors: | Garza-Garcia, A, Harris, R, Esposito, D, Driscoll, P.C, Rieping, W. | | Deposit date: | 2007-04-16 | | Release date: | 2008-02-26 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure and conformational plasticity of the N-terminal scavenger receptor cysteine-rich domain of human CD5
J.Mol.Biol., 378, 2008
|
|
