8AF6
 
 | | Room temperature SSX structure of GH11 xylanase from Nectria haematococca (4000 frames) | | Descriptor: | Endo-1,4-beta-xylanase | | Authors: | Oberthuer, D, Andaleeb, H, Betzel, C, Perbandt, M, Yefanov, O, Zielinski, K. | | Deposit date: | 2022-07-15 | | Release date: | 2022-11-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Rapid and efficient room-temperature serial synchrotron crystallography using the CFEL TapeDrive.
Iucrj, 9, 2022
|
|
6Y0H
 
 | | High resolution structure of GH11 xylanase from Nectria haematococca | | Descriptor: | Endo-1,4-beta-xylanase | | Authors: | Andaleeb, H, Betzel, C, Perbandt, M, Brognaro, H. | | Deposit date: | 2020-02-07 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | High-resolution crystal structure and biochemical characterization of a GH11 endoxylanase from Nectria haematococca.
Sci Rep, 10, 2020
|
|
2AYW
 
 | | Crystal Structure of the complex formed between trypsin and a designed synthetic highly potent inhibitor in the presence of benzamidine at 0.97 A resolution | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-[2-({[4-(DIAMINOMETHYL)PHENYL]AMINO}CARBONYL)-6-METHOXYPYRIDIN-3-YL]-5-{[(1-FORMYL-2,2-DIMETHYLPROPYL)AMINO]CARBONYL}BENZOIC ACID, BENZAMIDINE, ... | | Authors: | Sherawat, M, Kaur, P, Perbandt, M, Betzel, C, Slusarchyk, W.A, Bisacchi, G.S, Chang, C, Jacobson, B.L, Einspahr, H.M, Singh, T.P. | | Deposit date: | 2005-09-09 | | Release date: | 2006-01-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Structure of the complex of trypsin with a highly potent synthetic inhibitor at 0.97 A resolution.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
3C2X
 
 | | Crystal structure of peptidoglycan recognition protein at 1.8A resolution | | Descriptor: | GLYCEROL, L(+)-TARTARIC ACID, Peptidoglycan recognition protein, ... | | Authors: | Sharma, P, Singh, N, Sinha, M, Sharma, S, Perbandt, M, Betzel, C, Kaur, P, Srinivasan, A, Singh, T.P. | | Deposit date: | 2008-01-26 | | Release date: | 2008-03-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structure of the peptidoglycan recognition protein at 1.8 A resolution reveals dual strategy to combat infection through two independent functional homodimers
J.Mol.Biol., 378, 2008
|
|
3BU8
 
 | | Crystal Structure of TRF2 TRFH domain and TIN2 peptide complex | | Descriptor: | TERF1-interacting nuclear factor 2, Telomeric repeat-binding factor 2 | | Authors: | Chen, Y, Yang, Y, van Overbeek, M, Donigian, J.R, Baciu, P, de Lange, T, Lei, M. | | Deposit date: | 2008-01-02 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A shared docking motif in TRF1 and TRF2 used for differential recruitment of telomeric proteins.
Science, 319, 2008
|
|
3BUA
 
 | | Crystal Structure of TRF2 TRFH domain and APOLLO peptide complex | | Descriptor: | DNA cross-link repair 1B protein, Telomeric repeat-binding factor 2 | | Authors: | Chen, Y, Yang, Y, van Overbeek, M, Donigian, J.R, Baciu, P, de Lange, T, Lei, M. | | Deposit date: | 2008-01-02 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A shared docking motif in TRF1 and TRF2 used for differential recruitment of telomeric proteins.
Science, 319, 2008
|
|
3BQO
 
 | | Crystal Structure of TRF1 TRFH domain and TIN2 peptide complex | | Descriptor: | TERF1-interacting nuclear factor 2, Telomeric repeat-binding factor 1 | | Authors: | Chen, Y, Yang, Y, van Overbeek, M, Donigian, J.R, Baciu, P, de Lange, T, Lei, M. | | Deposit date: | 2007-12-20 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A shared docking motif in TRF1 and TRF2 used for differential recruitment of telomeric proteins.
Science, 319, 2008
|
|
7NEV
 
 | | Structure of the hemiacetal complex between the SARS-CoV-2 Main Protease and Leupeptin | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H.M, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashhour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Xavier, P.L, Ullah, N, Andaleeb, H, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Zaitsev-Doyle, J.J, Rogers, C, Gieseler, H, Melo, D, Monteiro, D.C.F, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schluenzen, F, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Sun, X, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2021-02-05 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7NFV
 
 | | Structure of SARS-CoV-2 Papain-like protease PLpro | | Descriptor: | CHLORIDE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Srinivasan, V, Gunther, S, Reinke, P, Werner, N, Falke, S, Brognaro, H, Ullah, N, Andaleeb, H, Perbandt, M, Alves Franca, B, Schwinzer, M, Wang, M, Sprenger, J, Lieske, J, Ginn, H, Lane, T.J, Yefanov, O, Gelisio, L, Koua, F, Saouane, S, Tolstikova, A, Groessler, M, Fleckenstein, H, Ewert, W, Trost, F, Lorenzen, K, Schubert, R, Han, H, Schmidt, C, Brings, L, Ehrt, C, Rarey, M, Galchenkova, M, Gevorkov, Y, Li, C, Perk, A, Awel, S, Hinrichs, W, Meents, A, Betzel, C. | | Deposit date: | 2021-02-07 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Antiviral activity of natural phenolic compounds in complex at an allosteric site of SARS-CoV-2 papain-like protease.
Commun Biol, 5, 2022
|
|
7OFU
 
 | | Structure of SARS-CoV-2 Papain-like protease PLpro in complex with 3, 4-Dihydroxybenzoic acid, methyl ester | | Descriptor: | CHLORIDE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Srinivasan, V, Ewert, W, Werner, N, Falke, S, Guenther, S, Reinke, P, Sprenger, J, Brognaro, H, Ullah, N, Andaleeb, H, Perbandt, M, Alves Franca, B, Schwinzer, M, Wang, M, Wolf, M, Lieske, J, Koua, F, Ginn, H, Lane, T.J, Yefanov, O, Gelisio, L, Hakanpaeae, J, Saouane, S, Tolstikova, A, Groessler, M, Fleckenstein, H, Trost, F, Lorenzen, K, Schubert, R, Han, H, Schmidt, C, Brings, L, Galchenkova, M, Gevorkov, Y, Li, C, Perk, A, Awel, S, Wahab, A, Choudary, I, Turk, D, Hinrichs, W, Chapman, H.N, Meents, A, Betzel, C. | | Deposit date: | 2021-05-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Antiviral activity of natural phenolic compounds in complex at an allosteric site of SARS-CoV-2 papain-like protease.
Commun Biol, 5, 2022
|
|
7OFS
 
 | | Structure of SARS-CoV-2 Papain-like protease PLpro in complex with 4-(2-hydroxyethyl)phenol | | Descriptor: | 4-(2-hydroxyethyl)phenol, CHLORIDE ION, GLYCEROL, ... | | Authors: | Srinivasan, V, Werner, N, Falke, S, Guenther, S, Reinke, P, Ewert, W, Sprenger, J, Koua, F, Brognaro, H, Ullah, N, Andaleeb, H, Perbandt, M, Alves Franca, B, Schwinzer, M, Wang, M, Lieske, J, Ginn, H, Lane, T.J, Yefanov, O, Gelisio, L, Hakanpaeae, J, Saouane, S, Tolstikova, A, Groessler, M, Fleckenstein, H, Trost, F, Wolf, M, Lorenzen, K, Schubert, R, Han, H, Schmidt, C, Brings, L, Galchenkova, M, Gevorkov, Y, Li, C, Perk, A, Awel, S, Wahab, A, Choudary, I, Turk, D, Hinrichs, W, Chapman, H.N, Meents, A, Betzel, C. | | Deposit date: | 2021-05-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Antiviral activity of natural phenolic compounds in complex at an allosteric site of SARS-CoV-2 papain-like protease.
Commun Biol, 5, 2022
|
|
7OFT
 
 | | Structure of SARS-CoV-2 Papain-like protease PLpro in complex with p-hydroxybenzaldehyde | | Descriptor: | CHLORIDE ION, P-HYDROXYBENZALDEHYDE, POTASSIUM ION, ... | | Authors: | Srinivasan, V, Werner, N, Falke, S, Guenther, S, Reinke, P, Brognaro, H, Ullah, N, Andaleeb, H, Perbandt, M, Alves Franca, B, Schwinzer, M, Wang, M, Ewert, W, Sprenger, J, Lieske, J, Koua, F, Ginn, H, Lane, T.J, Wolf, M, Yefanov, O, Gelisio, L, Saouane, S, Tolstikova, A, Groessler, M, Fleckenstein, H, Trost, F, Lorenzen, K, Schubert, R, Han, H, Schmidt, C, Brings, L, Galchenkova, M, Gevorkov, Y, Li, C, Perk, A, Awel, S, Wahab, A, Choudary, I, Turk, D, Hinrichs, W, Chapman, H.N, Meents, A, Betzel, C. | | Deposit date: | 2021-05-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Antiviral activity of natural phenolic compounds in complex at an allosteric site of SARS-CoV-2 papain-like protease.
Commun Biol, 5, 2022
|
|
2YVV
 
 | | Crystal structure of hyluranidase complexed with lactose at 2.6 A resolution reveals three specific sugar recognition sites | | Descriptor: | Hyaluronidase, phage associated, beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Mishra, P, Prem Kumar, R, Singh, N, Sharma, S, Kaur, P, Perbandt, M, Betzel, C, Bhakuni, V, Singh, T.P. | | Deposit date: | 2007-04-16 | | Release date: | 2007-05-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of hyluranidase complexed with lactose at 2.6 A resolution reveals three specific sugar recognition sites
To be Published
|
|
2YW0
 
 | | Crystal structure of hyluranidase trimer at 2.6 A resolution | | Descriptor: | Hyaluronidase, phage associated | | Authors: | Prem Kumar, R, Mishra, P, Singh, N, Perbandt, M, Kaur, P, Sharma, S, Betzel, C, Bhakuni, V, Singh, T.P. | | Deposit date: | 2007-04-18 | | Release date: | 2007-05-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Polysaccharide binding sites in hyaluronate lyase--crystal structures of native phage-encoded hyaluronate lyase and its complexes with ascorbic acid and lactose
Febs J., 276, 2009
|
|
2YX2
 
 | | Crystal structure of cloned trimeric hyluranidase from streptococcus pyogenes at 2.8 A resolution | | Descriptor: | Hyaluronidase, phage associated | | Authors: | Mishra, P, Prem Kumar, R, Bhakuni, V, Singh, N, Sharma, S, Kaur, P, Perbandt, M, Betzel, C, Singh, T.P. | | Deposit date: | 2007-04-23 | | Release date: | 2007-05-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of cloned trimeric hyluranidase from streptococcus pyogenes at 2.8 A resolution
To be Published
|
|
7OX8
 
 | | Target-bound SpCas9 complex with TRAC full RNA guide | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Donohoue, P, Pacesa, M, Lau, E, Vidal, B, Irby, M.J, Nyer, D.B, Rotstein, T, Banh, L, Toh, M.T, Gibson, J, Kohrs, B, Baek, K, Owen, A.L.G, Slorach, E.M, van Overbeek, M, Fuller, C.K, May, A.P, Jinek, M, Cameron, P. | | Deposit date: | 2021-06-22 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Conformational control of Cas9 by CRISPR hybrid RNA-DNA guides mitigates off-target activity in T cells.
Mol.Cell, 81, 2021
|
|
7OXA
 
 | | Target-bound SpCas9 complex with AAVS1 chimeric RNA-DNA guide | | Descriptor: | AAVS1 non-target DNA strand, AAVS1 target DNA strand, CRISPR-associated endonuclease Cas9/Csn1, ... | | Authors: | Donohoue, P, Pacesa, M, Lau, E, Vidal, B, Irby, M.J, Nyer, D.B, Rotstein, T, Banh, L, Toh, M.T, Gibson, J, Kohrs, B, Baek, K, Owen, A.L.G, Slorach, E.M, van Overbeek, M, Fuller, C.K, May, A.P, Jinek, M, Cameron, P. | | Deposit date: | 2021-06-22 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Conformational control of Cas9 by CRISPR hybrid RNA-DNA guides mitigates off-target activity in T cells.
Mol.Cell, 81, 2021
|
|
7OX7
 
 | | Target-bound SpCas9 complex with TRAC chimeric RNA-DNA guide | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Donohoue, P, Pacesa, M, Lau, E, Vidal, B, Irby, M.J, Nyer, D.B, Rotstein, T, Banh, L, Toh, M.T, Gibson, J, Kohrs, B, Baek, K, Owen, A.L.G, Slorach, E.M, van Overbeek, M, Fuller, C.K, May, A.P, Jinek, M, Cameron, P. | | Deposit date: | 2021-06-22 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Conformational control of Cas9 by CRISPR hybrid RNA-DNA guides mitigates off-target activity in T cells.
Mol.Cell, 81, 2021
|
|
6RFU
 
 | | In cellulo crystallization of Trypanosoma brucei IMP dehydrogenase enables the identification of ATP and GMP as genuine co-factors | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-MONOPHOSPHATE, Inosine-5'-monophosphate dehydrogenase | | Authors: | Nass, K, Redecke, L, Perbandt, M, Yefanov, O, Gabdulkhakov, A, Duszenko, M, Chapman, H.N, Betzel, C. | | Deposit date: | 2019-04-16 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | In cellulo crystallization of Trypanosoma brucei IMP dehydrogenase enables the identification of genuine co-factors.
Nat Commun, 11, 2020
|
|
4OW2
 
 | | YopM from Yersinia enterocolitica WA-314 | | Descriptor: | Yop effector YopM | | Authors: | Rumm, A, Perbandt, M, Aepfelbacher, M. | | Deposit date: | 2014-01-30 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Immunosuppressive Yersinia Effector YopM Binds DEAD Box Helicase DDX3 to Control Ribosomal S6 Kinase in the Nucleus of Host Cells.
PLoS Pathog., 12, 2016
|
|
5LH0
 
 | | Low dose Thaumatin - 0-40 ms. | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Schubert, R, Kapis, S, Heymann, M, Giquel, Y, Bourenkov, G, Schneider, T, Betzel, C, Perbandt, M. | | Deposit date: | 2016-07-08 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | A multicrystal diffraction data-collection approach for studying structural dynamics with millisecond temporal resolution.
IUCrJ, 3, 2016
|
|
5LH7
 
 | | High dose Thaumatin - 760-800 ms. | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Schubert, R, Kapis, S, Heymann, M, Giquel, Y, Bourenkov, G, Schneider, T, Betzel, C, Perbandt, M. | | Deposit date: | 2016-07-08 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | A multicrystal diffraction data-collection approach for studying structural dynamics with millisecond temporal resolution.
IUCrJ, 3, 2016
|
|
5LN0
 
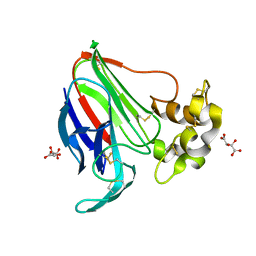 | | Low dose Thaumatin - 760-800 ms. | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Schubert, R, Kapis, S, Heymann, M, Giquel, Y, Bourenkov, G, Schneider, T, Betzel, C, Perbandt, M. | | Deposit date: | 2016-08-02 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A multicrystal diffraction data-collection approach for studying structural dynamics with millisecond temporal resolution.
IUCrJ, 3, 2016
|
|
8PCD
 
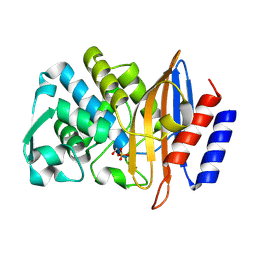 | | Structure of serine-beta-lactamase CTX-M-14 following the time-resolved active site binding of boric acid, 150 ms | | Descriptor: | BORATE ION, Beta-lactamase, SULFATE ION | | Authors: | Prester, A, Perbandt, M, Galchenkova, M, Oberthuer, D, Yefanov, O, Hinrichs, W, Rohde, H, Betzel, C. | | Deposit date: | 2023-06-11 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Time resolved crystallography of boric acid binding to the active site serine of the beta-lactamase CTX-M-14 and subsequent 1,2-diol esterification
To Be Published
|
|
8PCF
 
 | | Structure of serine-beta-lactamase CTX-M-14 following the time-resolved active site binding of boric acid, 500 ms | | Descriptor: | BORATE ION, Beta-lactamase, SULFATE ION | | Authors: | Prester, A, Perbandt, M, Galchenkova, M, Oberthuer, D, Yefanov, O, Hinrichs, W, Rohde, H, Betzel, C. | | Deposit date: | 2023-06-11 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Time resolved crystallography of boric acid binding to the active site serine of the beta-lactamase CTX-M-14 and subsequent 1,2-diol esterification
To Be Published
|
|
