2LEW
 
 | | Structural Plasticity of Paneth cell alpha-Defensins: Characterization of Salt-Bridge Deficient Analogues of Mouse Cryptdin-4 | | Descriptor: | Alpha-defensin 4 | | Authors: | Rosengren, K, Andersson, H.S, Haugaard-Kedstrom, L.M, Bengtsson, E, Daly, N.L, Craik, D.J. | | Deposit date: | 2011-06-24 | | Release date: | 2012-05-16 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | The alpha-defensin salt-bridge induces backbone stability to facilitate folding and confer proteolytic resistance.
Amino Acids, 43, 2012
|
|
2LR9
 
 | |
1MB6
 
 | |
5T4R
 
 | |
7YKM
 
 | |
6FWS
 
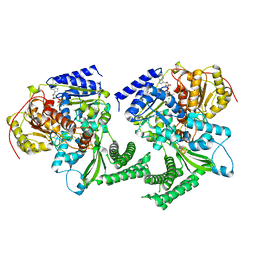 | | Structure of DinG in complex with ssDNA and ADPBeF | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase DinG, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Cheng, K, Wigley, D. | | Deposit date: | 2018-03-07 | | Release date: | 2018-12-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | DNA translocation mechanism of an XPD family helicase.
Elife, 7, 2018
|
|
6FWR
 
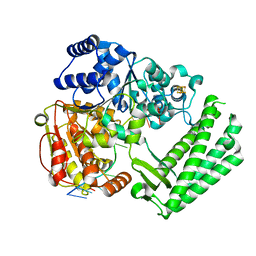 | | Structure of DinG in complex with ssDNA | | Descriptor: | ATP-dependent DNA helicase DinG, DNA (5'-D(*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), IRON/SULFUR CLUSTER | | Authors: | Cheng, K, Wigley, D.B. | | Deposit date: | 2018-03-07 | | Release date: | 2018-12-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | DNA translocation mechanism of an XPD family helicase.
Elife, 7, 2018
|
|
5MZP
 
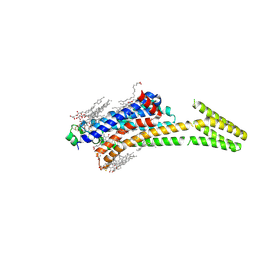 | | Crystal structure of stabilized A2A adenosine receptor A2AR-StaR2-bRIL in complex with caffeine at 2.1A resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, CAFFEINE, ... | | Authors: | Cheng, K.Y.R, Segala, E, Robertson, N, Deflorian, F, Dore, A.S, Errey, J.C, Fiez-Vandal, C, Marshall, F.H, Cooke, R.M. | | Deposit date: | 2017-02-01 | | Release date: | 2017-07-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of Human A1 and A2A Adenosine Receptors with Xanthines Reveal Determinants of Selectivity.
Structure, 25, 2017
|
|
7S3E
 
 | |
8IU7
 
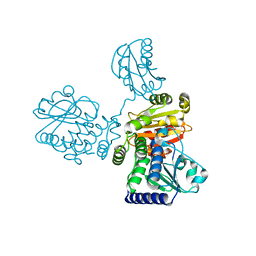 | |
8IOO
 
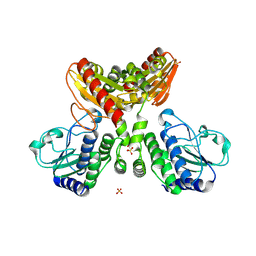 | |
6WPV
 
 | |
2LEY
 
 | | Solution structure of (R7G)-Crp4 | | Descriptor: | Alpha-defensin 4 | | Authors: | Rosengren, K, Andersson, H.S, Haugaard-Kedstrom, L.M, Bengtsson, E, Daly, N.L, Figueredo, S.M, Qu, X, Craik, D.J, Ouellette, A.J. | | Deposit date: | 2011-06-26 | | Release date: | 2012-05-16 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | The alpha-defensin salt-bridge induces backbone stability to facilitate folding and confer proteolytic resistance.
Amino Acids, 43, 2012
|
|
7L54
 
 | |
7LZL
 
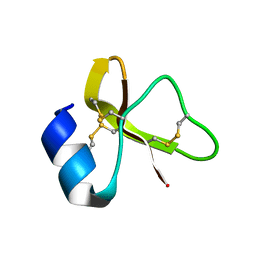 | |
7L0F
 
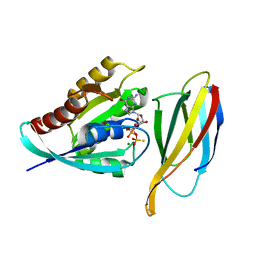 | | Monobody 12VC3 Bound to HRAS(WT) | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Teng, K.W, Hattori, T, Tsai, S, Koide, S. | | Deposit date: | 2020-12-11 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Selective and noncovalent targeting of RAS mutants for inhibition and degradation.
Nat Commun, 12, 2021
|
|
7L0G
 
 | | Monobody 12VC1 Bound to HRAS(G12C) | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Teng, K.W, Hattori, T, Tsai, S, Koide, S. | | Deposit date: | 2020-12-11 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Selective and noncovalent targeting of RAS mutants for inhibition and degradation.
Nat Commun, 12, 2021
|
|
3DEO
 
 | |
6SER
 
 | | Crystal structure of human STARD10 | | Descriptor: | DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, START domain-containing protein 10, ... | | Authors: | Cheng, K, Wigley, D.B. | | Deposit date: | 2019-07-30 | | Release date: | 2020-08-26 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | The crystal structure of human STARD10
To Be Published
|
|
6SJF
 
 | | Cryo-EM structure of the RecBCD Chi unrecognised complex | | Descriptor: | Forked DNA substrate, RecBCD enzyme subunit RecB, RecBCD enzyme subunit RecC, ... | | Authors: | Cheng, K, Wilkinson, M, Wigley, D.B. | | Deposit date: | 2019-08-13 | | Release date: | 2020-01-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | A conformational switch in response to Chi converts RecBCD from phage destruction to DNA repair.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6SJE
 
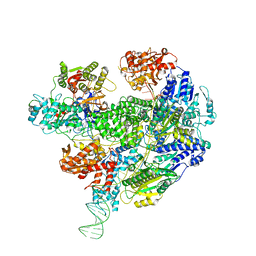 | | Cryo-EM structure of the RecBCD Chi partially-recognised complex | | Descriptor: | DNA fork substrate, RecBCD enzyme subunit RecB, RecBCD enzyme subunit RecC, ... | | Authors: | Cheng, K, Wilkinson, M, Wigley, D.B. | | Deposit date: | 2019-08-13 | | Release date: | 2020-01-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | A conformational switch in response to Chi converts RecBCD from phage destruction to DNA repair.
Nat.Struct.Mol.Biol., 27, 2020
|
|
2CCI
 
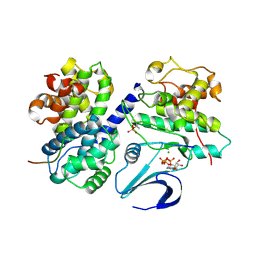 | | Crystal structure of phospho-CDK2 Cyclin A in complex with a peptide containing both the substrate and recruitment sites of CDC6 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 6 homolog, Cyclin-A2, ... | | Authors: | Cheng, K.Y, Noble, M.E.M, Skamnaki, V, Brown, N.R, Lowe, E.D, Kontogiannis, L, Shen, K, Cole, P.A, Siligardi, G, Johnson, L.N. | | Deposit date: | 2006-01-16 | | Release date: | 2006-05-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The role of the phospho-CDK2/cyclin A recruitment site in substrate recognition.
J. Biol. Chem., 281, 2006
|
|
2MP8
 
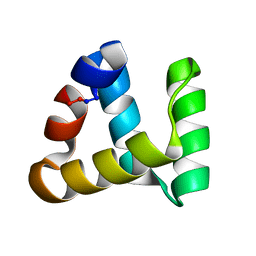 | | NMR structure of NKR-5-3B | | Descriptor: | NKR-5-3B | | Authors: | Rosengren, K.J, Craik, D.J. | | Deposit date: | 2014-05-13 | | Release date: | 2015-05-13 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Identification, Characterization, and Three-Dimensional Structure of the Novel Circular Bacteriocin, Enterocin NKR-5-3B, from Enterococcus faecium
Biochemistry, 54, 2015
|
|
2CCH
 
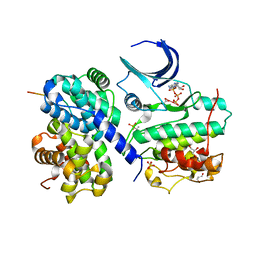 | | The crystal structure of CDK2 cyclin A in complex with a substrate peptide derived from CDC modified with a gamma-linked ATP analogue | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CELL DIVISION CONTROL PROTEIN 6 HOMOLOG, CELL DIVISION PROTEIN KINASE 2, ... | | Authors: | Cheng, K.Y, Noble, M.E.M, Skamnaki, V, Brown, N.R, Lowe, E.D, Kontogiannis, L, Shen, K, Cole, P.A, Siligardi, G, Johnson, L.N. | | Deposit date: | 2006-01-16 | | Release date: | 2006-05-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Role of the Phospho-Cdk2/Cyclin a Recruitment Site in Substrate Recognition
J.Biol.Chem., 281, 2006
|
|
6SJG
 
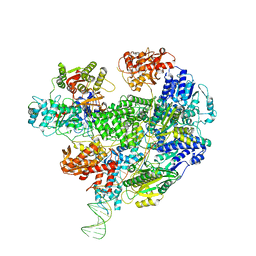 | | Cryo-EM structure of the RecBCD no Chi negative control complex | | Descriptor: | Forked DNA substrate, RecBCD enzyme subunit RecB, RecBCD enzyme subunit RecC, ... | | Authors: | Cheng, K, Wilkinson, M, Wigley, D.B. | | Deposit date: | 2019-08-13 | | Release date: | 2020-01-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A conformational switch in response to Chi converts RecBCD from phage destruction to DNA repair.
Nat.Struct.Mol.Biol., 27, 2020
|
|
