2OMQ
 
 | |
2ON9
 
 | |
2OMP
 
 | |
2ONA
 
 | |
3NHC
 
 | | GYMLGS segment 127-132 from human prion with M129 | | Descriptor: | Major prion protein | | Authors: | Apostol, M.I, Eisenberg, D. | | Deposit date: | 2010-06-14 | | Release date: | 2010-08-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystallographic studies of prion protein (PrP) segments suggest how structural changes encoded by polymorphism at residue 129 modulate susceptibility to human prion disease.
J.Biol.Chem., 285, 2010
|
|
3NI3
 
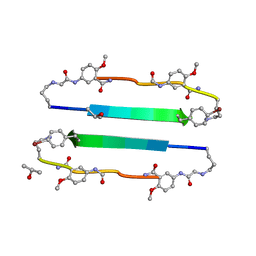 | | 54-Membered ring macrocyclic beta-sheet peptide | | Descriptor: | 54-membered ring macrocyclic beta-sheet peptide, ISOPROPYL ALCOHOL | | Authors: | Sawaya, M.R, Eisenberg, D, Nowick, J.S, Korman, T.P, Khakshoor, O. | | Deposit date: | 2010-06-15 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | X-ray crystallographic structure of an artificial beta-sheet dimer.
J.Am.Chem.Soc., 132, 2010
|
|
3NHD
 
 | | GYVLGS segment 127-132 from human prion with V129 | | Descriptor: | ACETIC ACID, Major prion protein | | Authors: | Apostol, M.I, Eisenberg, D. | | Deposit date: | 2010-06-14 | | Release date: | 2010-08-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystallographic studies of prion protein (PrP) segments suggest how structural changes encoded by polymorphism at residue 129 modulate susceptibility to human prion disease.
J.Biol.Chem., 285, 2010
|
|
3NVH
 
 | |
8DDH
 
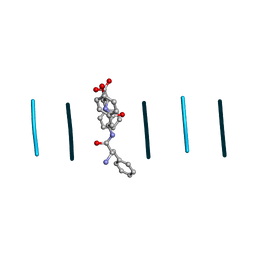 | |
7K3X
 
 | |
6B79
 
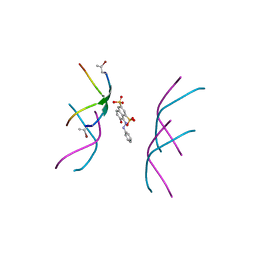 | |
6VW2
 
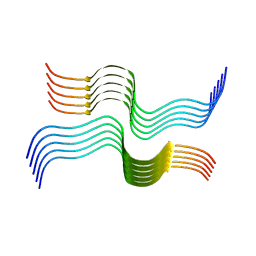 | | Cryo-EM structure of human islet amyloid polypeptide (hIAPP, or amylin) fibrils | | Descriptor: | Islet amyloid polypeptide (SUMO-tagged) | | Authors: | Cao, Q, Boyer, D.R, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2020-02-18 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure and inhibitor design of human IAPP (amylin) fibrils.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6WPQ
 
 | | GNYNVF from hnRNPA2-low complexity domain segment, residues 286-291, D290V variant | | Descriptor: | Heterogeneous nuclear ribonucleoprotein A2 | | Authors: | Lu, J, Cao, Q, Hughes, M.P, Sawaya, M.R, Boyer, D.R, Cascio, D, Eisenberg, D.S. | | Deposit date: | 2020-04-27 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | CryoEM structure of the low-complexity domain of hnRNPA2 and its conversion to pathogenic amyloid.
Nat Commun, 11, 2020
|
|
2QYG
 
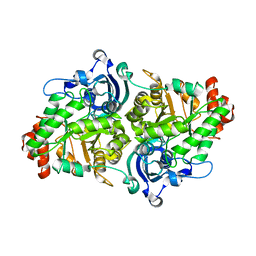 | | Crystal Structure of a RuBisCO-like Protein rlp2 from Rhodopseudomonas palustris | | Descriptor: | Ribulose bisphosphate carboxylase-like protein 2 | | Authors: | Li, H, Chan, S, Tabita, F.R, Eisenberg, D. | | Deposit date: | 2007-08-14 | | Release date: | 2007-09-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Function, structure, and evolution of the RubisCO-like proteins and their RubisCO homologs.
Microbiol.Mol.Biol.Rev., 71, 2007
|
|
8TER
 
 | | Tropomyosin-receptor kinase fused gene protein (TRK-fused gene protein; TFG) Low Complexity Domain (residues 237-327) P285L mutant, amyloid fiber | | Descriptor: | TRK-fused gene protein Low Complexity Domain P285L mutant | | Authors: | Rosenberg, G.M, Sawaya, M.R, Boyer, D.R, Ge, P, Abskharon, R, Eisenberg, D.S. | | Deposit date: | 2023-07-06 | | Release date: | 2023-12-20 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Fibril structures of TFG protein mutants validate the identification of TFG as a disease-related amyloid protein by the IMPAcT method.
Pnas Nexus, 2, 2023
|
|
8TEQ
 
 | | Tropomyosin-receptor kinase fused gene protein (TRK-fused gene protein; TFG) Low Complexity Domain (residues 237-327) G269V mutant, amyloid fiber | | Descriptor: | TRK-fused gene protein Low Complexity Domain G269V mutant | | Authors: | Rosenberg, G.M, Sawaya, M.R, Boyer, D.R, Ge, P, Abskharon, R, Eisenberg, D.S. | | Deposit date: | 2023-07-06 | | Release date: | 2023-12-20 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Fibril structures of TFG protein mutants validate the identification of TFG as a disease-related amyloid protein by the IMPAcT method.
Pnas Nexus, 2, 2023
|
|
7K3C
 
 | | SGMGGIT segment 58-64 from Keratin-8 | | Descriptor: | ETHANOL, SGMGGIT segment 58-64 from the low complexity domain of Keratin-8, TRIETHYLENE GLYCOL | | Authors: | Murray, K.A, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2020-09-11 | | Release date: | 2021-11-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Identifying amyloid-related diseases by mapping mutations in low-complexity protein domains to pathologies.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6AS9
 
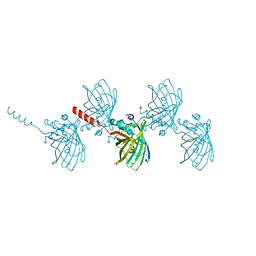 | | Filamentous Assembly of Green Fluorescent Protein Supported by a C-terminal fusion of 18-residues, viewed in space group P212121 form 2 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, Green fluorescent protein | | Authors: | Sawaya, M.R, Heller, D.M, McPartland, L, Hochschild, A, Eisenberg, D.S. | | Deposit date: | 2017-08-23 | | Release date: | 2018-05-30 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Atomic insights into the genesis of cellular filaments by globular proteins.
Nat. Struct. Mol. Biol., 25, 2018
|
|
8DDG
 
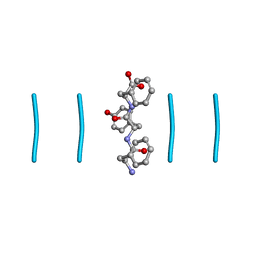 | |
6PQA
 
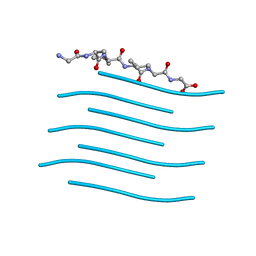 | |
8FQ7
 
 | | Nanobody with WIW inserted in CDR3 loop to Inhibit Growth of Alzheimer's Tau fibrils | | Descriptor: | GLYCEROL, Nanobody with WIW insert in CDR3 loop to target tau fibrils | | Authors: | Abskharon, R, Sawaya, M.R, Cascio, D.C, Eisenberg, D.S. | | Deposit date: | 2023-01-05 | | Release date: | 2023-10-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-based design of nanobodies that inhibit seeding of Alzheimer's patient-extracted tau fibrils.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7K3Y
 
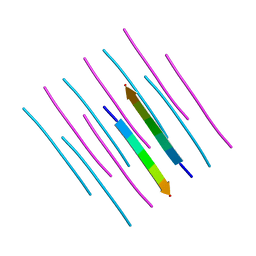 | | GGYAGAS segment 52-58 from Keratin-8 | | Descriptor: | GGYAGAS segment 52-58 from the low complexity domain of Keratin-8 | | Authors: | Murray, K.A, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2020-09-14 | | Release date: | 2021-11-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Identifying amyloid-related diseases by mapping mutations in low-complexity protein domains to pathologies.
Nat.Struct.Mol.Biol., 29, 2022
|
|
8DDF
 
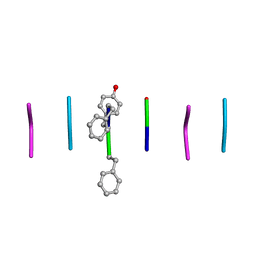 | | Quasi-racemic mixture of L-FWF and D-FYF peptide reveals rippled beta-sheet | | Descriptor: | 1,1,1,3,3,3-hexafluoropropan-2-ol, DPN-DTY-DPN, PHE-TRP-PHE | | Authors: | Sawaya, M.R, Hazari, A, Eisenberg, D.E. | | Deposit date: | 2022-06-18 | | Release date: | 2022-09-28 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The rippled beta-sheet layer configuration-a novel supramolecular architecture based on predictions by Pauling and Corey.
Chem Sci, 13, 2022
|
|
5VSG
 
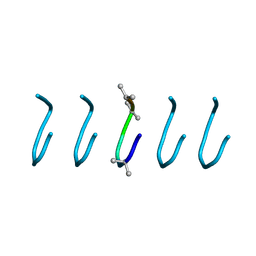 | | Fibrils of the super helical repeat peptide, SHR-FF, grown at elevated temperature | | Descriptor: | Super Helical Repeat Peptide SHR-FF | | Authors: | Mondal, S, Sawaya, M.R, Eisenberg, D.S, Gazit, E. | | Deposit date: | 2017-05-11 | | Release date: | 2018-06-27 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Transition of Metastable Cross-alpha Crystals into Cross-beta Fibrils by beta-Turn Flipping.
J.Am.Chem.Soc., 141, 2019
|
|
5E61
 
 | |
