6XOZ
 
 | |
1HJL
 
 | | Biochemical and Structural Analysis of the Molybdenum Cofactor Biosynthesis protein MobA | | Descriptor: | CITRIC ACID, LITHIUM ION, MOLYBDOPTERIN-GUANINE DINUCLEOTIDE BIOSYNTHESIS PROTEIN A | | Authors: | Guse, A, Stevenson, C.E.M, Kuper, J, Buchanan, G, Schwarz, G, Mendel, R.R, Lawson, D.M, Palmer, T. | | Deposit date: | 2003-02-27 | | Release date: | 2003-05-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemical and Structural Analysis of the Molybdenum Cofactor Biosynthesis Protein Moba
J.Biol.Chem., 278, 2003
|
|
1HJJ
 
 | | Biochemical and Structural Analysis of the Molybdenum Cofactor Biosynthesis protein MobA | | Descriptor: | CITRIC ACID, LITHIUM ION, MOLYBDOPTERIN-GUANINE DINUCLEOTIDE BIOSYNTHESIS PROTEIN A | | Authors: | Guse, A, Stevenson, C.E.M, Kuper, J, Buchanan, G, Schwarz, G, Mendel, R.R, Lawson, D.M, Palmer, T. | | Deposit date: | 2003-02-27 | | Release date: | 2003-05-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Biochemical and Structural Analysis of the Molybdenum Cofactor Biosynthesis Protein Moba
J.Biol.Chem., 278, 2003
|
|
8UBG
 
 | | DpHF19 filament | | Descriptor: | DpHF19,Green fluorescent protein (Fragment) | | Authors: | Lynch, E.M, Shen, H, Kollman, J.M, Baker, D. | | Deposit date: | 2023-09-22 | | Release date: | 2024-04-10 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | De novo design of pH-responsive self-assembling helical protein filaments.
Nat Nanotechnol, 19, 2024
|
|
8TQE
 
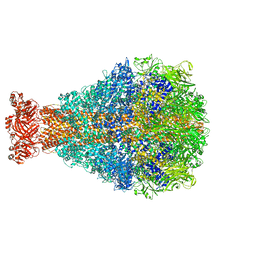 | | XptA2 wild type | | Descriptor: | XptA2 | | Authors: | Martin, C.L, Binshtein, E.M, Aller, S.G. | | Deposit date: | 2023-08-07 | | Release date: | 2023-09-27 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of the Insecticidal Toxin Complex Subunit XptA2 Highlight Roles for Flexible Domains.
Int J Mol Sci, 24, 2023
|
|
6XP2
 
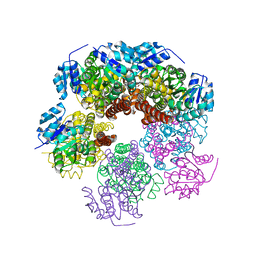 | |
5I2Y
 
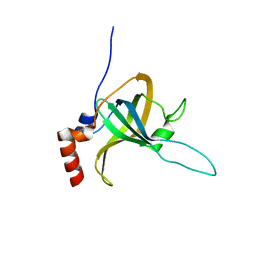 | | Crystal Structure of TPP1 K170A | | Descriptor: | Adrenocortical dysplasia protein homolog | | Authors: | Nandakumar, J, Smith, E.M. | | Deposit date: | 2016-02-09 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and functional consequences of a disease mutation in the telomere protein TPP1.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3CXC
 
 | | The structure of an enhanced oxazolidinone inhibitor bound to the 50S ribosomal subunit of H. marismortui | | Descriptor: | (3Z)-N-[(4E)-5-(4-{(5S)-5-[(acetylamino)methyl]-2-oxo-1,3-oxazolidin-3-yl}-2-fluorophenyl)pent-4-en-1-yl]-3-(4-methyl-2,6-dioxo-1,6-dihydropyrimidin-5(2H)-ylidene)propanamide, 23S RIBOSOMAL RNA, 5'-R(*CP*CP*A)-3', ... | | Authors: | Ippolito, J.A, Wang, D, Kanyo, Z.F, Duffy, E.M. | | Deposit date: | 2008-04-24 | | Release date: | 2009-04-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Design at the atomic level: design of biaryloxazolidinones as potent orally active antibiotics.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3DCQ
 
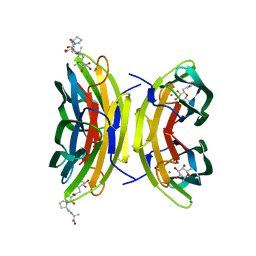 | | LECB (PA-LII) in complex with the synthetic ligand 2G0 | | Descriptor: | (2S)-1-[(2S)-6-amino-2-({[(2S,3S,4R,5S,6S)-3,4,5-trihydroxy-6-methyltetrahydro-2H-pyran-2-yl]acetyl}amino)hexanoyl]-N-[(1S)-1-carbamoyl-3-methylbutyl]pyrrolidine-2-carboxamide, CALCIUM ION, Fucose-binding lectin PA-IIL | | Authors: | Johansson, E.M, Crusz, S.A, Kolomiets, E, Buts, L, Kadam, R.U, Cacciarini, M, Bartels, K.M, Diggle, S.P, Camara, M, Williams, P, Loris, R, Nativi, C, Rosenau, F, Jaeger, K.E, Darbre, T, Reymond, J.L. | | Deposit date: | 2008-06-04 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Inhibition and dispersion of Pseudomonas aeruginosa biofilms by glycopeptide dendrimers targeting the fucose-specific lectin LecB.
Chem.Biol., 15, 2008
|
|
5V7X
 
 | | Crystal Structure of Myosin 1b residues 1-728 with bound sulfate and Calmodulin | | Descriptor: | Calmodulin-1, SULFATE ION, Unconventional myosin-Ib | | Authors: | Zwolak, A, Shuman, H, Dominguez, R, Ostap, E.M. | | Deposit date: | 2017-03-20 | | Release date: | 2018-02-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.141 Å) | | Cite: | High-resolution cryo-EM structures of actin-bound myosin states reveal the mechanism of myosin force sensing.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5I6Z
 
 | | X-ray structure of the ts2 human serotonin transporter | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 8B6 antibody, heavy chain, ... | | Authors: | Coleman, J.A, Green, E.M, Gouaux, E. | | Deposit date: | 2016-02-16 | | Release date: | 2016-04-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (4.53 Å) | | Cite: | X-ray structures and mechanism of the human serotonin transporter.
Nature, 532, 2016
|
|
5I73
 
 | | X-ray structure of the ts3 human serotonin transporter complexed with s-citalopram at the central and allosteric sites | | Descriptor: | (1S)-1-[3-(dimethylamino)propyl]-1-(4-fluorophenyl)-1,3-dihydro-2-benzofuran-5-carbonitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-AMINOHEXANOIC ACID, ... | | Authors: | Coleman, J.A, Green, E.M, Gouaux, E. | | Deposit date: | 2016-02-16 | | Release date: | 2016-04-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | X-ray structures and mechanism of the human serotonin transporter.
Nature, 532, 2016
|
|
6XP0
 
 | |
5W1F
 
 | | Crystal structure of Ni(II)- and Ca(II)-bound human calprotectin | | Descriptor: | CALCIUM ION, NICKEL (II) ION, Protein S100-A8, ... | | Authors: | Nakashige, T.G, Drennan, C.L, Nolan, E.M. | | Deposit date: | 2017-06-03 | | Release date: | 2017-06-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Nickel Sequestration by the Host-Defense Protein Human Calprotectin.
J. Am. Chem. Soc., 139, 2017
|
|
5IJC
 
 | | The crystal structure of mouse TLR4/MD-2/neoseptin-3 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lymphocyte antigen 96, ... | | Authors: | Wang, Y, Su, L, Morin, M.D, Jones, B.T, Whitby, L.R, Surakattula, M, Huang, H, Shi, H, Choi, J.H, Wang, K, Moresco, E.M, Berger, M, Zhan, X, Zhang, H, Boger, D.L, Beutler, B. | | Deposit date: | 2016-03-01 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | TLR4/MD-2 activation by a synthetic agonist with no similarity to LPS.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
8UAO
 
 | | DpHF18 filament | | Descriptor: | DpHF18 | | Authors: | Lynch, E.M, Shen, H, Kollman, J.M, Baker, D. | | Deposit date: | 2023-09-21 | | Release date: | 2024-04-10 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | De novo design of pH-responsive self-assembling helical protein filaments.
Nat Nanotechnol, 19, 2024
|
|
8UB3
 
 | | DpHF7 filament | | Descriptor: | DpHF7 filament | | Authors: | Lynch, E.M, Farrell, D, Shen, H, Kollman, J.M, DiMaio, F, Baker, D. | | Deposit date: | 2023-09-22 | | Release date: | 2024-04-10 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | De novo design of pH-responsive self-assembling helical protein filaments.
Nat Nanotechnol, 19, 2024
|
|
8TUQ
 
 | | STL Polyomavirus LTA NLS bound to importin alpha 2 | | Descriptor: | Importin subunit alpha-1, Large T antigen | | Authors: | Donnelly, C.M, Cross, E.M, Forwood, J.K, Alvisi, G. | | Deposit date: | 2023-08-17 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A functional and structural comparative analysis of large tumor antigens reveals evolution of different importin alpha-dependent nuclear localization signals.
Protein Sci., 33, 2024
|
|
8TUS
 
 | | WU Polyomavirus LTA NLS bound to importin alpha 2 | | Descriptor: | Importin subunit alpha-1, Large T antigen | | Authors: | Donnelly, C.M, Cross, E.M, Forwood, J.K, Alvisi, G. | | Deposit date: | 2023-08-17 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A functional and structural comparative analysis of large tumor antigens reveals evolution of different importin alpha-dependent nuclear localization signals.
Protein Sci., 33, 2024
|
|
8SPJ
 
 | |
5H83
 
 | | HETEROYOHIMBINE SYNTHASE HYS FROM CATHARANTHUS ROSEUS - APO FORM | | Descriptor: | ZINC ION, heteroyohimbine synthase HYS | | Authors: | Stavrinides, A, Tatsis, E.C, Caputi, L, Foureau, E, Stevenson, C.E.M, Lawson, D.M, Courdavault, V, O'Connor, S.E. | | Deposit date: | 2015-12-23 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural investigation of heteroyohimbine alkaloid synthesis reveals active site elements that control stereoselectivity.
Nat Commun, 7, 2016
|
|
8SXO
 
 | |
5IRV
 
 | | Human cytochrome P450 17A1 bound to inhibitor VT-464 | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Steroid 17-alpha-hydroxylase/17,20 lyase, VT-464 | | Authors: | Scott, E.E, Petrunak, E.M. | | Deposit date: | 2016-03-14 | | Release date: | 2017-03-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.098 Å) | | Cite: | Structural and Functional Evaluation of Clinically Relevant Inhibitors of Steroidogenic Cytochrome P450 17A1.
Drug Metab. Dispos., 45, 2017
|
|
5I66
 
 | | X-ray structure of the Fab fragment of 8B6, a murine monoclonal antibody specific for the human serotonin transporter | | Descriptor: | 8B6 antibody, heavy chain, light chain | | Authors: | Coleman, J.A, Green, E.M, Gouaux, E. | | Deposit date: | 2016-02-16 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.624 Å) | | Cite: | X-ray structures and mechanism of the human serotonin transporter.
Nature, 532, 2016
|
|
6XP1
 
 | |
