1CVX
 
 | | CRYSTAL STRUCTURE OF POLYAMIDE DIMER (IMPYHPPYBETADP)2 BOUND TO B-DNA DECAMER CCAGATCTGG | | Descriptor: | 5'-D(*CP*CP*AP*GP*AP*TP*CP*TP*GP*G)-3', HYDROXYPYRROLE-IMIDAZOLE-PYRROLE POLYAMIDE | | Authors: | Kielkopf, C.L, Bremer, R.E, White, S, Baird, E.E, Dervan, P.B, Rees, D.C. | | Deposit date: | 1999-08-24 | | Release date: | 2000-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural effects of DNA sequence on T.A recognition by hydroxypyrrole/pyrrole pairs in the minor groove.
J.Mol.Biol., 295, 2000
|
|
1BQ2
 
 | | E. COLI THYMIDYLATE SYNTHASE MUTANT N177A | | Descriptor: | PHOSPHATE ION, THYMIDYLATE SYNTHASE | | Authors: | Reyes, C.L, Sage, C.R, Rutenber, E.E, Finer-Moore, J.S, Stroud, R.M. | | Deposit date: | 1998-08-20 | | Release date: | 1999-04-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inactivity of N229A thymidylate synthase due to water-mediated effects: isolating a late stage in methyl transfer.
J.Mol.Biol., 284, 1998
|
|
1SNM
 
 | |
1R9K
 
 | | Representative solution structure of the catalytic domain of SopE2 | | Descriptor: | TypeIII-secreted protein effector: invasion-associated protein | | Authors: | Williams, C, Galyov, E.E, Bagby, S. | | Deposit date: | 2003-10-30 | | Release date: | 2004-09-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure, Backbone Dynamics, and Interaction with Cdc42 of Salmonella Guanine Nucleotide Exchange Factor SopE2(,).
Biochemistry, 43, 2004
|
|
1STY
 
 | |
1ENC
 
 | | CRYSTAL STRUCTURES OF THE BINARY CA2+ AND PDTP COMPLEXES AND THE TERNARY COMPLEX OF THE ASP 21->GLU MUTANT OF STAPHYLOCOCCAL NUCLEASE. IMPLICATIONS FOR CATALYSIS AND LIGAND BINDING | | Descriptor: | CALCIUM ION, STAPHYLOCOCCAL NUCLEASE, THYMIDINE-3',5'-DIPHOSPHATE | | Authors: | Libson, A, Gittis, A, Lattman, E.E. | | Deposit date: | 1994-02-14 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of the binary Ca2+ and pdTp complexes and the ternary complex of the Asp21-->Glu mutant of staphylococcal nuclease. Implications for catalysis and ligand binding.
Biochemistry, 33, 1994
|
|
1ENA
 
 | |
7Z3Z
 
 | | Locked Wuhan SARS-CoV2 Prefusion Spike ectodomain with lipid bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, STEARIC ACID, ... | | Authors: | Duyvesteyn, H.M.E, Carrique, L, Ren, J, Stuart, D.I, Fry, E.E. | | Deposit date: | 2022-03-03 | | Release date: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The SARS-CoV-2 Spike harbours a lipid binding pocket which modulates stability of the prefusion trimer
bioRxiv, 2020
|
|
6QJS
 
 | | Crystal structure of a DNA dodecamer containing a tetramethylpiperidinoxyl (nitroxide) spin label | | Descriptor: | COBALT HEXAMMINE(III), DNA (5'-D(P*GP*CP*AP*AP*AP*TP*TP*(S6M)P*GP*CP*G)-3') | | Authors: | Hardwick, J.S, Haugland, M.M, Ptchelkine, D, Brown, T, Anderson, E.E. | | Deposit date: | 2019-01-24 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 2'-Alkynyl spin-labelling is a minimally perturbing tool for DNA structural analysis.
Nucleic Acids Res., 48, 2020
|
|
6QJR
 
 | | Crystal structure of a DNA dodecamer containing a tetramethylpyrrolinoxyl (nitroxide) spin label | | Descriptor: | DNA (5'-D(*CP*GP*CP*AP*AP*AP*AP*AP*AP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*TP*TP*TP*TP*TP*(5NO)P*GP*CP*G)-3') | | Authors: | Hardwick, J.S, Haugland, M.M, Ptchelkine, D, Brown, T, Anderson, E.E. | | Deposit date: | 2019-01-24 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | 2'-Alkynyl spin-labelling is a minimally perturbing tool for DNA structural analysis.
Nucleic Acids Res., 48, 2020
|
|
1CMY
 
 | | THE MUTATION BETA99 ASP-TYR STABILIZES Y-A NEW, COMPOSITE QUATERNARY STATE OF HUMAN HEMOGLOBIN | | Descriptor: | HEMOGLOBIN YPSILANTI (CARBONMONOXY) (ALPHA CHAIN), HEMOGLOBIN YPSILANTI (CARBONMONOXY) (BETA CHAIN), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Smith, F.R, Lattman, E.E, Carter Junior, C.W. | | Deposit date: | 1992-09-18 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The mutation beta 99 Asp-Tyr stabilizes Y--a new, composite quaternary state of human hemoglobin.
Proteins, 10, 1991
|
|
1GEN
 
 | | C-TERMINAL DOMAIN OF GELATINASE A | | Descriptor: | CALCIUM ION, CHLORIDE ION, GELATINASE A, ... | | Authors: | Libson, A.M, Gittis, A.G, Collier, I.E, Marmer, B.L, Goldberg, G.G, Lattman, E.E. | | Deposit date: | 1995-07-19 | | Release date: | 1996-08-17 | | Last modified: | 2018-03-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of the haemopexin-like C-terminal domain of gelatinase A.
Nat.Struct.Biol., 2, 1995
|
|
1CVY
 
 | | CRYSTAL STRUCTURE OF POLYAMIDE DIMER (IMPYPYPYBETADP)2 BOUND TO CCAGATCTGG | | Descriptor: | 5'-D(*CP*CP*AP*GP*AP*TP*CP*TP*GP*G)-3', IMIDAZOLE-PYRROLE POLYAMIDE | | Authors: | Kielkopf, C.L, Bremer, R.E, White, S, Baird, E.E, Dervan, P.B, Rees, D.C. | | Deposit date: | 1999-08-24 | | Release date: | 2000-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural effects of DNA sequence on T.A recognition by hydroxypyrrole/pyrrole pairs in the minor groove.
J.Mol.Biol., 295, 2000
|
|
7LZ3
 
 | | Computational design of constitutively active cGAS | | Descriptor: | Cyclic GMP-AMP synthase, GLYCEROL, ZINC ION | | Authors: | Dowling, Q, Volkman, H.E, Gray, E.E, Ovchinnikov, S, Cambier, S, Bera, A.K, Bick, M, Kang, A, Stetson, D.B, King, N.P. | | Deposit date: | 2021-03-08 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Computational design of constitutively active cGAS.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7LYX
 
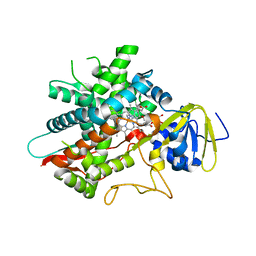 | | Crystal structure of human CYP8B1 in complex with (S)-tioconazole | | Descriptor: | (S)-Tioconazole, 7-alpha-hydroxycholest-4-en-3-one 12-alpha-hydroxylase, GLYCEROL, ... | | Authors: | Liu, J, Scott, E.E. | | Deposit date: | 2021-03-08 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure and characterization of human cytochrome P450 8B1 supports future drug design for nonalcoholic fatty liver disease and diabetes.
J.Biol.Chem., 298, 2022
|
|
7LU8
 
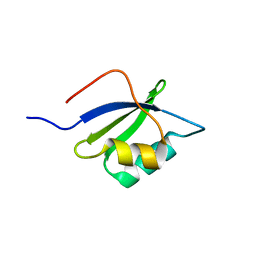 | |
7M8V
 
 | | Human CYP11B2 in complex with LCI699 | | Descriptor: | 4-[(4R,5R)-6,7-dihydro-5H-pyrrolo[1,2-c]imidazol-5-yl]-3-fluorobenzonitrile, Cytochrome P450 11B2, mitochondrial, ... | | Authors: | Scott, E.E, Brixius-Anderko, S. | | Deposit date: | 2021-03-30 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Aldosterone Synthase Structure With Cushing Disease Drug LCI699 Highlights Avenues for Selective CYP11B Drug Design.
Hypertension, 78, 2021
|
|
7M8H
 
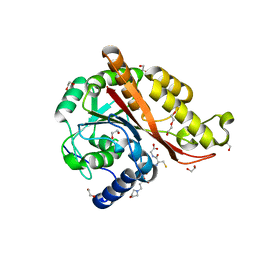 | | Structure of Memo1 C244S metal binding site mutant at 1.75A | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Boniecki, M.T, Uhlemann, E.E, Dmitriev, O.Y. | | Deposit date: | 2021-03-29 | | Release date: | 2022-04-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | MEMO1 binds iron and modulates iron homeostasis in cancer cells.
Elife, 13, 2024
|
|
7N9U
 
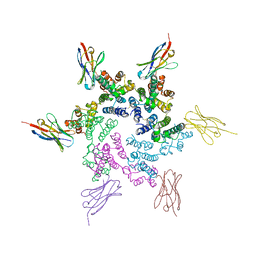 | | CA-targeting nanobody is a tool for studying HIV-1 capsid lattice interactions | | Descriptor: | Capsid protein, Nanobody | | Authors: | Gerber, E.E, Digianantonio, K.M, Tripler, T.N, Smaga, S.S, Summers, B.J, Xiong, Y. | | Deposit date: | 2021-06-18 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | CA-targeting nanobody is a tool for studying HIV-1 capsid lattice interactions
To Be Published
|
|
7N9X
 
 | | CA-targeting nanobody is a tool for studying HIV-1 capsid lattice interactions | | Descriptor: | Capsid protein, Nanobody, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Gerber, E.E, Digianantonio, K.M, Tripler, T.N, Smaga, S.S, Summers, B.J, Xiong, Y. | | Deposit date: | 2021-06-18 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.511 Å) | | Cite: | CA-targeting nanobody is a tool for studying HIV-1 capsid lattice interactions
To Be Published
|
|
7M8I
 
 | | Human CYP11B2 and human adrenodoxin in complex with fadrozole | | Descriptor: | 4-[(5R)-5,6,7,8-tetrahydroimidazo[1,5-a]pyridin-5-yl]benzonitrile, Adrenodoxin, Cytochrome P450 11B2, ... | | Authors: | Scott, E.E, Brixius-Anderko, S. | | Deposit date: | 2021-03-29 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Structural and functional insights into aldosterone synthase interaction with its redox partner protein adrenodoxin.
J.Biol.Chem., 296, 2021
|
|
7AY8
 
 | | NMR solution structure of Tbo-IT2 | | Descriptor: | Tbo-IT2 | | Authors: | Mineev, K.S, Kornilov, F.D, Lushpa, V.A, Korolkova, Y.A, Maleeva, E.E, Andreev, Y.A, Kozlov, S.A, Arseniev, A.S. | | Deposit date: | 2020-11-11 | | Release date: | 2021-03-03 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | New Insectotoxin from Tibellus Oblongus Spider Venom Presents Novel Adaptation of ICK Fold.
Toxins, 13, 2021
|
|
1SND
 
 | | STAPHYLOCOCCAL NUCLEASE DIMER CONTAINING A DELETION OF RESIDUES 114-119 COMPLEXED WITH CALCIUM CHLORIDE AND THE COMPETITIVE INHIBITOR DEOXYTHYMIDINE-3',5'-DIPHOSPHATE | | Descriptor: | STAPHYLOCOCCAL NUCLEASE DIMER | | Authors: | Green, S.M, Gittis, A.G, Meeker, A.K, Lattman, E.E. | | Deposit date: | 1996-08-23 | | Release date: | 1997-04-21 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | One-step evolution of a dimer from a monomeric protein.
Nat.Struct.Biol., 2, 1995
|
|
1RRE
 
 | | Crystal structure of E.coli Lon proteolytic domain | | Descriptor: | ATP-dependent protease La, SULFATE ION | | Authors: | Botos, I, Melnikov, E.E, Cherry, S, Tropea, J.E, Khalatova, A.G, Rasulova, F, Dauter, Z, Maurizi, M.R, Rotanova, T.V, Wlodawer, A, Gustchina, A. | | Deposit date: | 2003-12-08 | | Release date: | 2004-02-03 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The catalytic domain of Escherichia coli Lon protease has a unique fold and a Ser-Lys dyad in the active site
J.Biol.Chem., 279, 2004
|
|
1REI
 
 | | THE MOLECULAR STRUCTURE OF A DIMER COMPOSED OF THE VARIABLE PORTIONS OF THE BENCE-JONES PROTEIN REI REFINED AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | BENCE-JONES PROTEIN REI (LIGHT CHAIN) | | Authors: | Epp, O, Lattman, E.E, Colman, P, Fehlhammer, H, Bode, W, Schiffer, M, Huber, R, Palm, W. | | Deposit date: | 1976-03-17 | | Release date: | 1976-05-19 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The molecular structure of a dimer composed of the variable portions of the Bence-Jones protein REI refined at 2.0-A resolution.
Biochemistry, 14, 1975
|
|
