6X8M
 
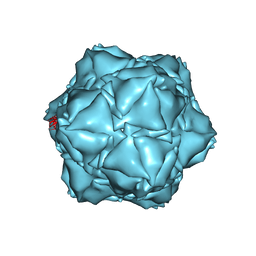 | | CryoEM structure of the holo-SrpI encapsulin complex from Synechococcus elongatus PCC 7942 | | Descriptor: | Protein SrpI | | Authors: | LaFrance, B.J, Nichols, R.J, Phillips, N.R, Oltrogge, L.M, Valentin-Alvarado, L.E, Bischoff, A.J, Savage, D.F, Nogales, E. | | Deposit date: | 2020-06-01 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Discovery and characterization of a novel family of prokaryotic nanocompartments involved in sulfur metabolism.
Elife, 10, 2021
|
|
8GA1
 
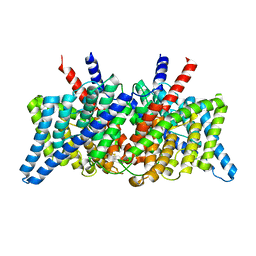 | | CLC-ec1 R230C/L249C/C85A at pH 4.5 100mM Cl Swap | | Descriptor: | CHLORIDE ION, H(+)/Cl(-) exchange transporter ClcA | | Authors: | Fortea, E, Lee, S, Argyos, Y, Chadda, R, Ciftci, D, Huysmans, G, Robertson, J.L, Boudker, O, Accardi, A. | | Deposit date: | 2023-02-22 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis of pH-dependent activation in a CLC transporter.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8GA5
 
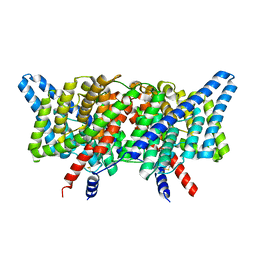 | | CLC-ec1 L25C/A450C/C85A at pH 4.5 100mM Cl Intermediate | | Descriptor: | CHLORIDE ION, H(+)/Cl(-) exchange transporter ClcA | | Authors: | Fortea, E, Lee, S, Argyos, Y, Chadda, R, Ciftci, D, Huysmans, G, Robertson, J.L, Boudker, O, Accardi, A. | | Deposit date: | 2023-02-22 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis of pH-dependent activation in a CLC transporter.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6FA3
 
 | | Antibody derived (Abd-6) small molecule binding to KRAS. | | Descriptor: | DIMETHYL SULFOXIDE, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Quevedo, C.E, Cruz-Migoni, A, Ehebauer, M.T, Carr, S.B, Phillips, S.E.V, Rabbitts, T.H. | | Deposit date: | 2017-12-15 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Small molecule inhibitors of RAS-effector protein interactions derived using an intracellular antibody fragment.
Nat Commun, 9, 2018
|
|
6OAZ
 
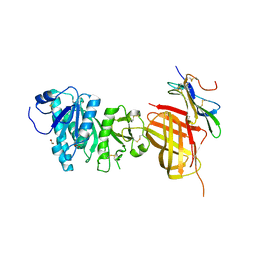 | | Apo Structure of WT Lipoprotein Lipase in Complex with GPIHBP1 Mutant N78D N82D produced in HEK293-F cells | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Arora, R, Horton, P.A, Benson, T.E, Romanowski, M.J. | | Deposit date: | 2019-03-19 | | Release date: | 2019-05-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Structure of lipoprotein lipase in complex with GPIHBP1.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
8SVK
 
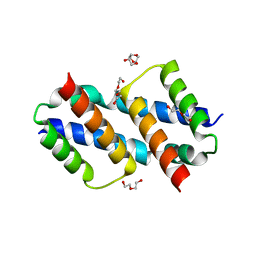 | | Crystal structure of Bax D71N core domain BH3-groove dimer | | Descriptor: | Apoptosis regulator BAX, DI(HYDROXYETHYL)ETHER, SULFATE ION, ... | | Authors: | Miller, M.S, Czabotar, P.E, Colman, P.M. | | Deposit date: | 2023-05-16 | | Release date: | 2023-12-27 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Sequence differences between BAX and BAK core domains manifest as differences in their interactions with lipids.
Febs J., 291, 2024
|
|
4U9C
 
 | |
8K6Y
 
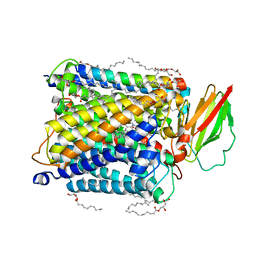 | | Serial femtosecond crystallography structure of photo dissociated CO from ba3- type cytochrome c oxidase determined by extrapolation method | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CARBON MONOXIDE, COPPER (II) ION, ... | | Authors: | Safari, C, Ghosh, S, Andersson, R, Johannesson, J, Donoso, A.V, Zoric, D, Sandelin, E, Iwata, S, Neutze, R, Branden, G. | | Deposit date: | 2023-07-25 | | Release date: | 2023-11-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Time-resolved serial crystallography to track the dynamics of carbon monoxide in the active site of cytochrome c oxidase.
Sci Adv, 9, 2023
|
|
5C3H
 
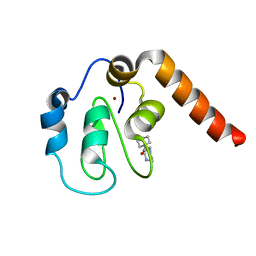 | | Fragment-Based Drug Discovery Targeting Inhibitor of Apoptosis Proteins: Compound 1 | | Descriptor: | 4-[2-oxo-2-(piperidin-1-yl)ethyl]piperazin-1-ium, E3 ubiquitin-protein ligase XIAP, ZINC ION | | Authors: | Chessari, G, Buck, I.M, Day, J.E.H, Day, P.J, Iqbal, A, Johnson, C.N, Lewis, E.J, Martins, V, Miller, D, Reader, M, Rees, D.C, Rich, S.J, Tamanini, E, Vitorino, M, Ward, G.A, Williams, P.A, Williams, G, Wilsher, N.E, Woolford, A.J.-A. | | Deposit date: | 2015-06-17 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Fragment-Based Drug Discovery Targeting Inhibitor of Apoptosis Proteins: Discovery of a Non-Alanine Lead Series with Dual Activity Against cIAP1 and XIAP.
J.Med.Chem., 58, 2015
|
|
5LRE
 
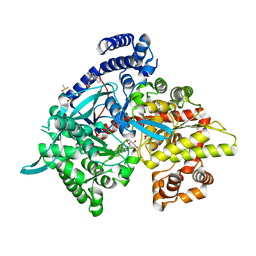 | | Crystal structure of Glycogen Phosphorylase b in complex with KS382 | | Descriptor: | (2~{R},3~{S},4~{R},5~{R},6~{S})-2-(hydroxymethyl)-6-(3-naphthalen-2-yl-1~{H}-1,2,4-triazol-5-yl)oxane-3,4,5-triol, DIMETHYL SULFOXIDE, Glycogen phosphorylase, ... | | Authors: | Kantsadi, A.L, Stravodimos, G.A, Kyriakis, E, Chatzileontiadou, D.S.M, Leonidas, D.D. | | Deposit date: | 2016-08-18 | | Release date: | 2017-05-31 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthetic, enzyme kinetic, and protein crystallographic studies of C-beta-d-glucopyranosyl pyrroles and imidazoles reveal and explain low nanomolar inhibition of human liver glycogen phosphorylase.
Eur J Med Chem, 123, 2016
|
|
8GKS
 
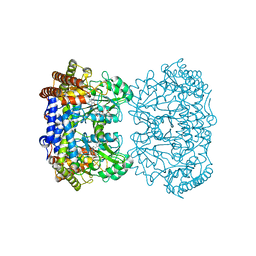 | | Human mitochondrial serine hydroxymethyltransferase (SHMT2) in complex with PLP, glycine and AGF291 inhibitor | | Descriptor: | GLYCINE, N-{4-[3-(2-amino-4-oxo-3,4-dihydro-5H-pyrrolo[3,2-d]pyrimidin-5-yl)propyl]benzoyl}-L-glutamic acid, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Katinas, J.M, Dann III, C.E. | | Deposit date: | 2023-03-20 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural Characterization of 5-Substituted Pyrrolo[3,2- d ]pyrimidine Antifolate Inhibitors in Complex with Human Serine Hydroxymethyl Transferase 2.
Biochemistry, 2024
|
|
8GKY
 
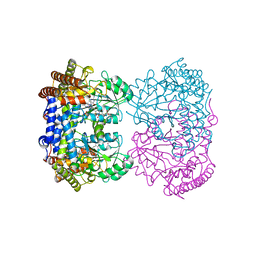 | | Human mitochondrial serine hydroxymethyltransferase (SHMT2) Y105F in complex with PLP, glycine and AGF359 inhibitor | | Descriptor: | GLYCINE, N-{4-[3-(2-amino-4-oxo-3,4-dihydro-5H-pyrrolo[3,2-d]pyrimidin-5-yl)propyl]-2-fluorobenzoyl}-L-glutamic acid, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Katinas, J.M, Dann III, C.E. | | Deposit date: | 2023-03-20 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structural Characterization of 5-Substituted Pyrrolo[3,2- d ]pyrimidine Antifolate Inhibitors in Complex with Human Serine Hydroxymethyl Transferase 2.
Biochemistry, 2024
|
|
5C7A
 
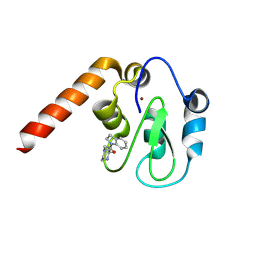 | | Fragment-Based Drug Discovery Targeting Inhibitor of Apoptosis Proteins: Compound 7 | | Descriptor: | (2R)-4-[2-(2,3-dihydro-1H-indol-1-yl)-2-oxoethyl]-2-methylpiperazin-1-ium, E3 ubiquitin-protein ligase XIAP, ZINC ION | | Authors: | Chessari, G, Buck, I.M, Day, J.E.H, Day, P.J, Iqbal, A, Johnson, C.N, Lewis, E.J, Martins, V, Miller, D, Reader, M, Rees, D.C, Rich, S.J, Tamanini, E, Vitorino, M, Ward, G.A, Williams, P.A, Williams, G, Wilsher, N.E, Woolford, A.J.-A. | | Deposit date: | 2015-06-24 | | Release date: | 2015-08-12 | | Last modified: | 2015-09-09 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Fragment-Based Drug Discovery Targeting Inhibitor of Apoptosis Proteins: Discovery of a Non-Alanine Lead Series with Dual Activity Against cIAP1 and XIAP.
J.Med.Chem., 58, 2015
|
|
4TWY
 
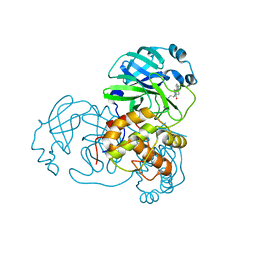 | | Structure of SARS-3CL protease complex with a phenylbenzoyl (S,R)-N-decalin type inhibitor | | Descriptor: | (2S)-2-({[(3S,4aR,8aS)-2-(biphenyl-4-ylcarbonyl)decahydroisoquinolin-3-yl]methyl}amino)-3-(1H-imidazol-5-yl)propanal, 3C-like proteinase | | Authors: | Akaji, K, Teruya, K, Shimamoto, Y, Sanjho, A, Yamashita, E, Nakagawa, A. | | Deposit date: | 2014-07-02 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Fused-ring structure of decahydroisoquinolin as a novel scaffold for SARS 3CL protease inhibitors
Bioorg.Med.Chem., 23, 2015
|
|
6XE3
 
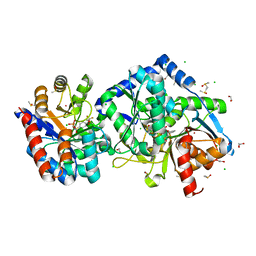 | | Salmonella typhimurium Tryptophan Synthase beta-S377A mutant in complex with inhibitor F9 at the enzyme alpha-site, cesium ion at the metal coordination site and carbanion III E(C3) at the enzyme beta-site. | | Descriptor: | (2E)-2-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)imino]-3-[(2-hydroxyphenyl)amino]propanoic acid, 1,2-ETHANEDIOL, 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, ... | | Authors: | Hilario, E, Dunn, M.F, Mueller, L.J, Fan, L. | | Deposit date: | 2020-06-11 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Salmonella typhimurium Tryptophan Synthase beta-S377A mutant in complex with inhibitor F9 at the enzyme alpha-site, cesium ion at the metal coordination site and carbanion III E(C3) at the enzyme beta-site.
To be Published
|
|
6ONS
 
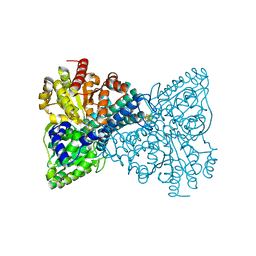 | | Crystal structure of Desulfovibrio vulgaris carbon monoxide dehydrogenase with the D-cluster ligating cysteines mutated to alanines, coexpressed with CooC, as-isolated | | Descriptor: | Carbon monoxide dehydrogenase, FE(4)-NI(1)-S(4) CLUSTER, IRON/SULFUR CLUSTER | | Authors: | Cohen, S.E, Wittenborn, E.C, Drennan, C.L. | | Deposit date: | 2019-04-22 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.485 Å) | | Cite: | Structural insight into metallocofactor maturation in carbon monoxide dehydrogenase.
J.Biol.Chem., 294, 2019
|
|
3GRS
 
 | |
4U1Z
 
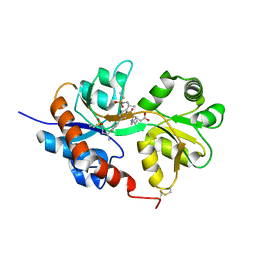 | | GluA2flip sLBD complexed with kainate and (R,R)-2b crystal form D | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Glutamate receptor 2,Glutamate receptor 2, N,N'-[biphenyl-4,4'-diyldi(2R)propane-2,1-diyl]dipropane-2-sulfonamide | | Authors: | Chen, L, Gouaux, E. | | Deposit date: | 2014-07-16 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9401 Å) | | Cite: | Structure and Dynamics of AMPA Receptor GluA2 in Resting, Pre-Open, and Desensitized States.
Cell, 158, 2014
|
|
6OQY
 
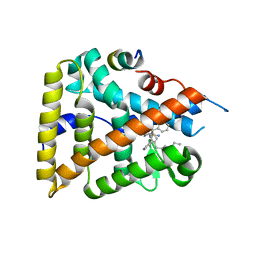 | | Human LRH-1 bound to the agonist 6N and a fragment of the Tif2 coregulator | | Descriptor: | N-[(1S,3aR,6aR)-5-hexyl-4-phenyl-3a-(1-phenylethenyl)-1,2,3,3a,6,6a-hexahydropentalen-1-yl]sulfuric diamide, Nuclear receptor coactivator 2, Nuclear receptor subfamily 5 group A member 2 | | Authors: | Mays, S.G, Ortlund, E.A. | | Deposit date: | 2019-04-29 | | Release date: | 2019-08-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Development of the First Low Nanomolar Liver Receptor Homolog-1 Agonist through Structure-guided Design.
J.Med.Chem., 62, 2019
|
|
4U22
 
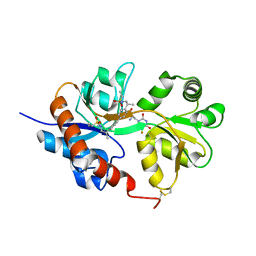 | | GluA2flip sLBD complexed with FW and (R,R)-2b crystal form D | | Descriptor: | 2-AMINO-3-(5-FLUORO-2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, Glutamate receptor 2, N,N'-[biphenyl-4,4'-diyldi(2R)propane-2,1-diyl]dipropane-2-sulfonamide | | Authors: | Chen, L, Gouaux, E. | | Deposit date: | 2014-07-16 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4409 Å) | | Cite: | Structure and Dynamics of AMPA Receptor GluA2 in Resting, Pre-Open, and Desensitized States.
Cell, 158, 2014
|
|
4U23
 
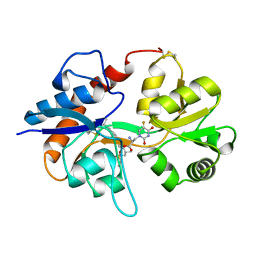 | | GluA2flip sLBD complexed with FW and (R,R)-2b crystal form F | | Descriptor: | 2-AMINO-3-(5-FLUORO-2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, Glutamate receptor 2,Glutamate receptor 2, N,N'-[biphenyl-4,4'-diyldi(2R)propane-2,1-diyl]dipropane-2-sulfonamide | | Authors: | Chen, L, Gouaux, E. | | Deposit date: | 2014-07-16 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6734 Å) | | Cite: | Structure and Dynamics of AMPA Receptor GluA2 in Resting, Pre-Open, and Desensitized States.
Cell, 158, 2014
|
|
4U5N
 
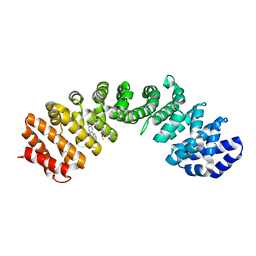 | | IMPORTIN-ALPHA MINOR NLS SITE INHIBITOR | | Descriptor: | Importin subunit alpha-1, N~2~-[4-(pyridin-3-yl)benzyl]-L-lysyl-N-[(1R,2S,3R)-1-{[(2R)-1-amino-1-oxo-3-phenylpropan-2-yl]amino}-1,3-dihydroxybutan-2-yl]glycinamide | | Authors: | Stewart, M, Valkov, E, Holvey, R.S. | | Deposit date: | 2014-07-25 | | Release date: | 2015-05-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Selective Targeting of the TPX2 Site of Importin-alpha Using Fragment-Based Ligand Design.
Chemmedchem, 10, 2015
|
|
4U5S
 
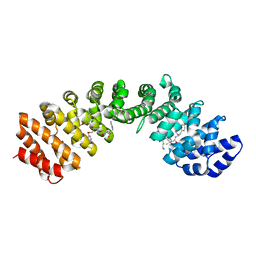 | | IMPORTIN-ALPHA MINOR NLS SITE INHIBITOR | | Descriptor: | Importin subunit alpha-1, N-[(2S)-2-[(N~2~-acetyl-D-lysyl)amino]-3-(pyridin-3-ylmethoxy)propyl]-L-allothreonyl-D-phenylalaninamide | | Authors: | Stewart, M, Valkov, E, Holvey, R.S. | | Deposit date: | 2014-07-25 | | Release date: | 2015-05-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Selective Targeting of the TPX2 Site of Importin-alpha Using Fragment-Based Ligand Design.
Chemmedchem, 10, 2015
|
|
8CIK
 
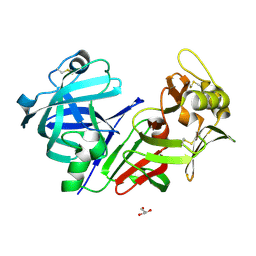 | | Altai wapiti (Cervus elaphus sibiricus) chymosin at 2.2 A resolution | | Descriptor: | Chymosin, GLYCEROL | | Authors: | Diusenova, S.E, Shevtsov, M.B, Borshchevskiy, V.I, Belenkaya, S.V, Kolybalov, D.S, Arkhipov, S.G, Volosnikova, E.A, Elchaninov, V.V, Shcherbakov, D.N. | | Deposit date: | 2023-02-09 | | Release date: | 2023-03-08 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Altai wapiti (Cervus elaphus sibiricus) chymosin at 2.2 A resolution
To Be Published
|
|
8GKT
 
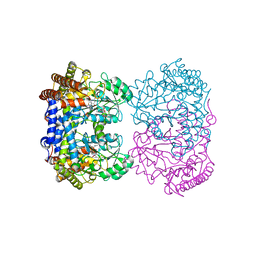 | | Human mitochondrial serine hydroxymethyltransferase (SHMT2) in complex with PLP, glycine and AGF320 inhibitor | | Descriptor: | N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], N-{5-[5-(2-amino-4-oxo-3,4-dihydro-5H-pyrrolo[3,2-d]pyrimidin-5-yl)pentyl]thiophene-2-carbonyl}-L-glutamic acid, Serine hydroxymethyltransferase, ... | | Authors: | Katinas, J.M, Dann III, C.E. | | Deposit date: | 2023-03-20 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structural Characterization of 5-Substituted Pyrrolo[3,2- d ]pyrimidine Antifolate Inhibitors in Complex with Human Serine Hydroxymethyl Transferase 2.
Biochemistry, 2024
|
|
