2R9M
 
 | | Cathepsin S complexed with Compound 15 | | 分子名称: | Cathepsin S, N-[(1S)-2-[(4-cyano-1-methylpiperidin-4-yl)amino]-1-(cyclohexylmethyl)-2-oxoethyl]morpholine-4-carboxamide | | 著者 | Ward, Y.D, Emmanuel, M.J, Thomson, D.S, Liu, W, Bekkali, Y, Frye, L.L, Girardot, M, Morwick, T, Young, E.R.R, Zindell, R, Hrapchak, M, DeTuri, M, White, A, Crane, K.M, White, D.M, Wang, Y, Hao, M.-H, Grygon, C.A, Labadia, M.E, Wildeson, J, Freeman, D, Nelson, R, Capolino, A, Peterson, J.D, Raymond, E.L, Brown, M.L, Spero, D.M. | | 登録日 | 2007-09-13 | | 公開日 | 2007-12-18 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Design and Synthesis of Reversible Inhibitors of Cathepsin S: alpha,alpha-Disubstitution at the P1 Residue Provides Potent Inhibitors in Cellular Assays and In Vivo Models of Antigen Presentation
To be Published
|
|
2DPF
 
 | | Crystal Structure of curculin1 homodimer | | 分子名称: | Curculin, SULFATE ION | | 著者 | Kurimoto, E, Suzuki, M, Amemiya, E, Yamaguchi, Y, Nirasawa, S, Shimba, N, Xu, N, Kashiwagi, T, Kawai, M, Suzuki, E, Kato, K. | | 登録日 | 2006-05-11 | | 公開日 | 2007-05-15 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Curculin Exhibits Sweet-tasting and Taste-modifying Activities through Its Distinct Molecular Surfaces.
J.Biol.Chem., 282, 2007
|
|
6M7L
 
 | | Complex of OxyA with the X-domain from GPA biosynthesis | | 分子名称: | PROTOPORPHYRIN IX CONTAINING FE, Putative cytochrome P450 hydroxylase, Putative non-ribosomal peptide synthetase | | 著者 | Greule, A, Izore, T, Tailhades, J, Peschke, M, Schoppet, M, Ahmed, I, Kulik, A, Adamek, M, Ziemert, N, De Voss, J, Stegmann, E, Cryle, M.J. | | 登録日 | 2018-08-20 | | 公開日 | 2019-05-22 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.648297 Å) | | 主引用文献 | Kistamicin biosynthesis reveals the biosynthetic requirements for production of highly crosslinked glycopeptide antibiotics.
Nat Commun, 10, 2019
|
|
1AGG
 
 | | THE SOLUTION STRUCTURE OF OMEGA-AGA-IVB, A P-TYPE CALCIUM CHANNEL ANTAGONIST FROM THE VENOM OF AGELENOPSIS APERTA | | 分子名称: | OMEGA-AGATOXIN-IVB | | 著者 | Reily, M.D, Thanabal, V, Adams, M.E. | | 登録日 | 1995-11-03 | | 公開日 | 1996-03-08 | | 最終更新日 | 2024-06-05 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of omega-Aga-IVB, a P-type calcium channel antagonist from venom of the funnel web spider, Agelenopsis aperta.
J.Biomol.NMR, 5, 1995
|
|
1T8W
 
 | |
1J53
 
 | | Structure of the N-terminal Exonuclease Domain of the Epsilon Subunit of E.coli DNA Polymerase III at pH 8.5 | | 分子名称: | 1,2-ETHANEDIOL, DNA polymerase III, epsilon chain, ... | | 著者 | Hamdan, S, Carr, P.D, Brown, S.E, Ollis, D.L, Dixon, N.E. | | 登録日 | 2002-01-22 | | 公開日 | 2002-10-16 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural Basis for Proofreading during Replication of the Escherichia coli Chromosome
Structure, 10, 2002
|
|
6ZOI
 
 | | A lid blocking mechanism of a cone snail toxin revealed at the atomic level | | 分子名称: | Conknunitzin-C3 mutante | | 著者 | Saikia, C, Altman-Gueta, H, Dym, O, Frolow, F, Gurevitz, M, Gordon, D, Reuveny, E, Karbat, I. | | 登録日 | 2020-07-07 | | 公開日 | 2021-07-14 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.798 Å) | | 主引用文献 | A lid blocking mechanism of a cone snail toxin revealed at the atomic level
To Be Published
|
|
140L
 
 | |
3LPE
 
 | | Crystal structure of Spt4/5NGN heterodimer complex from Methanococcus jannaschii | | 分子名称: | DNA-directed RNA polymerase subunit E'', Putative transcription antitermination protein nusG, ZINC ION | | 著者 | Hirtreiter, A, Damsma, G.E, Cheung, A.C.M, Klose, D, Grohmann, D, Vojnic, E, Martin, A.C.R, Cramer, P, Werner, F. | | 登録日 | 2010-02-05 | | 公開日 | 2010-03-09 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Spt4/5 stimulates transcription elongation through the RNA polymerase clamp coiled-coil motif.
Nucleic Acids Res., 38, 2010
|
|
6I9K
 
 | | Crystal structure of Jumping Spider Rhodopsin-1 bound to 9-cis retinal | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Kumopsin1, RETINAL | | 著者 | Varma, N, Mutt, E, Muehle, J, Panneels, V, Terakita, A, Deupi, X, Nogly, P, Schertler, F.X.G, Lesca, E. | | 登録日 | 2018-11-23 | | 公開日 | 2019-07-03 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.145 Å) | | 主引用文献 | Crystal structure of jumping spider rhodopsin-1 as a light sensitive GPCR.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
147L
 
 | |
141L
 
 | |
143L
 
 | |
142L
 
 | |
6IOL
 
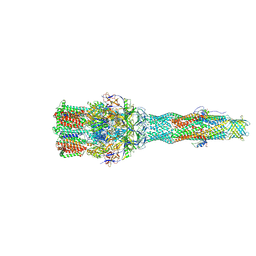 | | Cryo-EM structure of multidrug efflux pump MexAB-OprM (60 degree state) | | 分子名称: | Multidrug resistance protein MexA, Multidrug resistance protein MexB, Outer membrane protein OprM | | 著者 | Tsutsumi, K, Yonehara, R, Nakagawa, A, Yamashita, E. | | 登録日 | 2018-10-30 | | 公開日 | 2019-04-03 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.76 Å) | | 主引用文献 | Structures of the wild-type MexAB-OprM tripartite pump reveal its complex formation and drug efflux mechanism.
Nat Commun, 10, 2019
|
|
4F8X
 
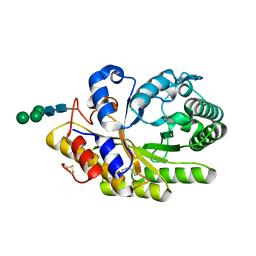 | | Penicillium canescens endo-1,4-beta-xylanase XylE | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Endo-1,4-beta-xylanase, beta-D-mannopyranose-(1-3)-[beta-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Polyakov, K.M, Trofimov, A.A, Fedorova, T.V, Koroleva, O.V, Maisuradze, I.G, Chulkin, A.M, Vavilova, E.A, Benevolenskii, S.V. | | 登録日 | 2012-05-18 | | 公開日 | 2012-05-30 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.47 Å) | | 主引用文献 |
|
|
6UEH
 
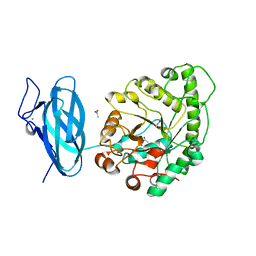 | | Crystal structure of a ruminal GH26 endo-beta-1,4-mannanase | | 分子名称: | ACETATE ION, CALCIUM ION, Cow rumen GH26 endo-mannanase | | 著者 | Mandelli, F, Morais, M.A.B, Lima, E.A, Persinoti, G.F, Murakami, M.T. | | 登録日 | 2019-09-21 | | 公開日 | 2020-03-11 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.849 Å) | | 主引用文献 | Spatially remote motifs cooperatively affect substrate preference of a ruminal GH26-type endo-beta-1,4-mannanase.
J.Biol.Chem., 295, 2020
|
|
7Z2P
 
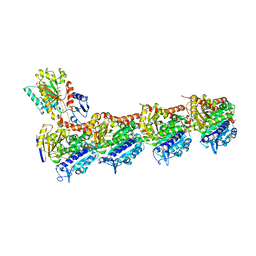 | | Tubulin-nocodazole complex | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Prota, A.E, Muehlethaler, T, Cavalli, A, Steinmetz, M.O. | | 登録日 | 2022-02-28 | | 公開日 | 2022-08-31 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Novel fragment-derived colchicine-site binders as microtubule-destabilizing agents.
Eur.J.Med.Chem., 241, 2022
|
|
7Z2N
 
 | | Tubulin-18-complex | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | 著者 | Muehlethaler, T, Prota, A.E, Cavalli, A, Steinmetz, M.O. | | 登録日 | 2022-02-28 | | 公開日 | 2022-08-31 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.17 Å) | | 主引用文献 | Novel fragment-derived colchicine-site binders as microtubule-destabilizing agents.
Eur.J.Med.Chem., 241, 2022
|
|
8BYA
 
 | | Cryo-EM structure of SKP1-SKP2-CKS1-CDK2-CyclinA-p27KIP1 Complex | | 分子名称: | Cyclin-A2, Cyclin-dependent kinase 2, Cyclin-dependent kinase inhibitor 1B, ... | | 著者 | Rowland, R.J, Salamina, M, Endicott, J.A, Noble, M.E. | | 登録日 | 2022-12-12 | | 公開日 | 2023-06-28 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (3.38 Å) | | 主引用文献 | Cryo-EM structure of SKP1-SKP2-CKS1 in complex with CDK2-cyclin A-p27KIP1.
Sci Rep, 13, 2023
|
|
6UIG
 
 | |
1A1H
 
 | | QGSR (ZIF268 VARIANT) ZINC FINGER-DNA COMPLEX (GCAC SITE) | | 分子名称: | DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*CP*AP*C)-3'), DNA (5'-D(*TP*GP*TP*GP*CP*CP*CP*AP*CP*GP*C)-3'), QGSR ZINC FINGER PEPTIDE, ... | | 著者 | Elrod-Erickson, M, Benson, T.E, Pabo, C.O. | | 登録日 | 1997-12-10 | | 公開日 | 1998-06-10 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | High-resolution structures of variant Zif268-DNA complexes: implications for understanding zinc finger-DNA recognition.
Structure, 6, 1998
|
|
1A1R
 
 | | HCV NS3 PROTEASE DOMAIN:NS4A PEPTIDE COMPLEX | | 分子名称: | NS3 PROTEIN, NS4A PROTEIN, ZINC ION | | 著者 | Kim, J.L, Morgenstern, K.A, Lin, C, Fox, T, Dwyer, M.D, Landro, J.A, Chambers, S.P, Markland, W, Lepre, C.A, O'Malley, E.T, Harbeson, S.L, Rice, C.M, Murcko, M.A, Caron, P.R, Thomson, J.A. | | 登録日 | 1997-12-15 | | 公開日 | 1998-06-17 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of the hepatitis C virus NS3 protease domain complexed with a synthetic NS4A cofactor peptide.
Cell(Cambridge,Mass.), 87, 1996
|
|
8C0F
 
 | | Tubulin-PTC596 complex | | 分子名称: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-fluoranyl-2-(6-fluoranyl-2-methyl-benzimidazol-1-yl)-~{N}4-[4-(trifluoromethyl)phenyl]pyrimidine-4,6-diamine, ... | | 著者 | Prota, A.E, Muehlethaler, T, Weetall, M, Steinmetz, M.O. | | 登録日 | 2022-12-16 | | 公開日 | 2022-12-28 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.1005 Å) | | 主引用文献 | Preclinical and Early Clinical Development of PTC596, a Novel Small-Molecule Tubulin-Binding Agent
Mol Cancer Ther, 20, 2021
|
|
2R9O
 
 | | Cathepsin S complexed with Compound 8 | | 分子名称: | Cathepsin S, N-[(1S)-2-{[(1R)-2-(benzyloxy)-1-cyano-1-methylethyl]amino}-1-(cyclohexylmethyl)-2-oxoethyl]morpholine-4-carboxamide | | 著者 | Ward, Y.D, Emmanuel, M.J, Thomson, D.S, Liu, W, Bekkali, Y, Frye, L.L, Girardot, M, Morwick, T, Young, E.R.R, Zindell, R, Hrapchak, M, DeTuri, M, White, A, Crane, K.M, White, D.M, Wang, Y, Hao, M.-H, Grygon, C.A, Labadia, M.E, Wildeson, J, Freeman, D, Nelson, R, Capolino, A, Peterson, J.D, Raymond, E.L, Brown, M.L, Spero, D.M. | | 登録日 | 2007-09-13 | | 公開日 | 2007-12-18 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Design and Synthesis of Reversible Inhibitors of Cathepsin S: alpha,alpha-Disubstitution at the P1 Residue Provides Potent Inhibitors in Cellular Assays and In Vivo Models of Antigen Presentation
to be published
|
|
