9BJ1
 
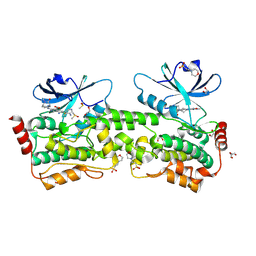 | | Crystal structure of inhibitor GNE-6893 bound to HPK1 | | Descriptor: | (4S,5R,7R,11aP)-10-{[(3R)-3-hydroxy-1-methyl-2-oxopyrrolidin-3-yl]ethynyl}-N~3~-methyl-6,7-dihydro-5H-5,7-methanoimidazo[2,1-a][2]benzazepine-2,3-dicarboxamide, (9S)-2-{[(6P)-8-amino-6-(5-amino-4-methylpyridin-3-yl)-7-fluoroisoquinolin-3-yl]amino}-6-methyl-5,6-dihydro-4H-pyrazolo[1,5-d][1,4]diazepin-7(8H)-one, 1,2-ETHANEDIOL, ... | | Authors: | Kiefer, J.R, Tellis, J.C, Chan, B.K, Wang, W, Wu, P, Choo, E.F, Heffron, T.P, Wei, B, Siu, M. | | Deposit date: | 2024-04-24 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Discovery of GNE-6893, a Potent, Selective, Orally Bioavailable Small Molecule Inhibitor of HPK1.
Acs Med.Chem.Lett., 15, 2024
|
|
2FGG
 
 | | Crystal Structure of Rv2632c | | Descriptor: | Hypothetical protein Rv2632c/MT2708 | | Authors: | Yu, M, Bursey, E.H, Radhakannan, T, Segelke, B.W, Lekin, T, Toppani, D, Kim, C.Y, Kaviratne, T, Woodruff, T, Terwilliger, T.C, Hung, L.W, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2005-12-21 | | Release date: | 2006-02-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Rv2632c
To be Published
|
|
2FJ6
 
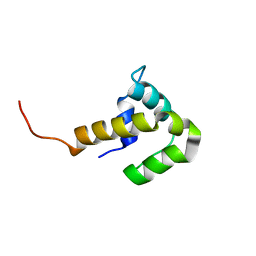 | | Solution NMR structure of the UPF0346 protein yozE from Bacillus subtilis. Northeast Structural Genomics target SR391. | | Descriptor: | Hypothetical UPF0346 protein yozE | | Authors: | Rossi, P, Acton, T.B, Cunningham, K.E, Ma, L.C, Shetty, K, Swapna, G.V.T, Xiao, R, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-12-31 | | Release date: | 2006-02-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the UPF0346 protein yozE from Bacillus subtilis. Northeast Structural Genomics target SR391.
To be Published
|
|
2F28
 
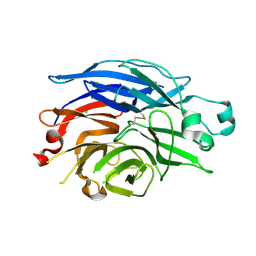 | | Crystal Structure of the Human Sialidase Neu2 Q116E Mutant | | Descriptor: | CHLORIDE ION, Sialidase 2 | | Authors: | Chavas, L.M.G, Kato, R, Fusi, P, Tringali, C, Venerando, B, Tettamanti, G, Monti, E, Wakatsuki, S. | | Deposit date: | 2005-11-15 | | Release date: | 2006-11-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal Structure of the Human Sialidase Neu2 Q116E Mutant
To be Published
|
|
2F77
 
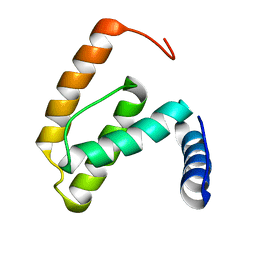 | | Solution structure of the R55F mutant of M-PMV matrix protein (p10) | | Descriptor: | Core protein p10 | | Authors: | Vlach, J, Lipov, J, Veverka, V, Lang, J, Srb, P, Rumlova, M, Hunter, E, Ruml, T, Hrabal, R. | | Deposit date: | 2005-11-30 | | Release date: | 2006-12-05 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | D-retrovirus morphogenetic switch driven by the targeting signal accessibility to Tctex-1 of dynein.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2FOI
 
 | | Synthesis, Biological Activity, and X-Ray Crystal Structural Analysis of Diaryl Ether Inhibitors of Malarial Enoyl ACP Reductase. | | Descriptor: | 4-(2,4-DICHLOROPHENOXY)-2'-METHYLBIPHENYL-3-OL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, enoyl-acyl carrier reductase | | Authors: | Freundlich, J.S, Shieh, H, Anderson, J.W, Kuo, M, Yu, M, Valderramos, J, Karagyozov, L, Tsai, H, Lucumi, E, Jacobs Jr, W.R, Schiehser, G.A, Jacobus, D.P, Fidock, D.A, Sacchettini, J.C. | | Deposit date: | 2006-01-13 | | Release date: | 2007-01-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray Structural Analysis of Plasmodium falciparum Enoyl Acyl Carrier Protein Reductase as a Pathway toward the Optimization of Triclosan Antimalarial Efficacy.
J.Biol.Chem., 282, 2007
|
|
2FWQ
 
 | |
2FUB
 
 | | Crystal structure of urate oxidase at 140 MPa | | Descriptor: | 8-AZAXANTHINE, CYSTEINE, Uricase | | Authors: | Colloc'h, N, Girard, E, Fourme, R. | | Deposit date: | 2006-01-26 | | Release date: | 2006-02-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | High pressure macromolecular crystallography: The 140-MPa crystal structure at 2.3 A resolution of urate oxidase, a 135-kDa tetrameric assembly
Biochim.Biophys.Acta, 1764, 2006
|
|
2FZF
 
 | | Hypothetical Protein Pfu-1136390-001 From Pyrococcus furiosus | | Descriptor: | hypothetical protein | | Authors: | Fu, Z.-Q, Liu, Z.-J, Lee, D, Kelley, L, Chen, L, Tempel, W, Shah, N, Horanyi, P, Lee, H.S, Habel, J, Dillard, B.D, Nguyen, D, Chang, S.-H, Zhang, H, Chang, J, Sugar, F.J, Poole, F.L, Jenney Jr, F.E, Adams, M.W.W, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2006-02-09 | | Release date: | 2006-02-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Hypothetical Protein Pfu-1136390-001 From Pyrococcus furiosus
To be published
|
|
2FV8
 
 | | The crystal structure of RhoB in the GDP-bound state | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Rho-related GTP-binding protein RhoB | | Authors: | Turnbull, A.P, Soundararajan, M, Smee, C, Johansson, C, Schoch, G, Gorrec, F, Bray, J, Papagrigoriou, E, von Delft, F, Weigelt, J, Edwards, A, Arrowsmith, C, Sundstrom, M, Doyle, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-01-30 | | Release date: | 2006-02-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of RhoB in the GDP-bound state
To be Published
|
|
2FW3
 
 | | Crystal structure of rat carnitine palmitoyltransferase 2 in complex with antidiabetic drug ST1326 | | Descriptor: | (3R)-3-{[(TETRADECYLAMINO)CARBONYL]AMINO}-4-(TRIMETHYLAMMONIO)BUTANOATE, Carnitine O-palmitoyltransferase II, mitochondrial | | Authors: | Rufer, A.C, Thoma, R, Benz, J, Stihle, M, Gsell, B, De Roo, E, Banner, D.W, Mueller, F, Chomienne, O, Hennig, M. | | Deposit date: | 2006-02-01 | | Release date: | 2007-02-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of carnitine palmitoyltransferase 2 and implications for diabetes treatment
Structure, 14, 2006
|
|
2FR9
 
 | | NMR structure of the alpha-conotoxin GI (SER12)-benzoylphenylalanine derivative | | Descriptor: | Alpha-conotoxin GI | | Authors: | Pashkov, V.S, Maslennikov, I.V, Kasheverov, I.E, Zhmak, M.N, Utkin, Y.N, Tsetlin, V.I, Arseniev, A.S. | | Deposit date: | 2006-01-19 | | Release date: | 2006-05-30 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Alpha-Conotoxin GI benzoylphenylalanine derivatives. (1)H-NMR structures and photoaffinity labeling of the Torpedo californica nicotinic acetylcholine receptor.
Febs J., 273, 2006
|
|
2FZU
 
 | | Reduced enolate chromophore intermediate for GFP variant | | Descriptor: | 1,2-ETHANEDIOL, Green fluorescent protein, MAGNESIUM ION | | Authors: | Barondeau, D.P, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2006-02-10 | | Release date: | 2006-03-14 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural evidence for an enolate intermediate in GFP fluorophore biosynthesis.
J.Am.Chem.Soc., 128, 2006
|
|
2G9J
 
 | | Complex of TM1a(1-14)Zip with TM9a(251-284): a model for the polymerization domain ("overlap region") of tropomyosin, Northeast Structural Genomics Target OR9 | | Descriptor: | Tropomyosin 1 alpha chain, Tropomyosin 1 alpha chain/General control protein GCN4 | | Authors: | Greenfield, N.J, Huang, Y.J, Swapna, G.V.T, Bhattacharya, A, Singh, A, Montelione, G.T, Hitchcock-DeGregori, S.E, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-03-06 | | Release date: | 2006-11-07 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of the Junction between Tropomyosin Molecules: Implications for Actin Binding and Regulation.
J.Mol.Biol., 364, 2006
|
|
4V7M
 
 | | The structures of Capreomycin bound to the 70S ribosome. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Stanley, R.E, Blaha, G. | | Deposit date: | 2009-11-12 | | Release date: | 2014-07-09 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | The structures of the anti-tuberculosis antibiotics viomycin and capreomycin bound to the 70S ribosome.
Nat.Struct.Mol.Biol., 17, 2010
|
|
4WBX
 
 | | Conserved hypothetical protein PF1771 from Pyrococcus furiosus solved by sulfur SAD using Swiss Light Source data | | Descriptor: | 2-keto acid:ferredoxin oxidoreductase subunit alpha | | Authors: | Weinert, T, Waltersperger, S, Olieric, V, Panepucci, E, Chen, L, Rose, J.P, Wang, M, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2014-09-04 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
2QFG
 
 | | Solution Structure of the N-terminal SCR-1/5 fragment of Complement Factor H. | | Descriptor: | Complement factor H | | Authors: | Okemefuna, A.I, Gilbert, H.E, Griggs, K.M, Ormsby, R.J, Gordon, D.L, Perkins, S.J. | | Deposit date: | 2007-06-27 | | Release date: | 2007-09-25 | | Last modified: | 2024-02-21 | | Method: | SOLUTION SCATTERING | | Cite: | The regulatory SCR-1/5 and cell surface-binding SCR-16/20 fragments of factor H reveal partially folded-back solution structures and different self-associative properties.
J.Mol.Biol., 375, 2008
|
|
4UYU
 
 | |
4UZ7
 
 | |
2QSV
 
 | | Crystal structure of protein of unknown function from Porphyromonas gingivalis W83 | | Descriptor: | SODIUM ION, SULFATE ION, Uncharacterized protein | | Authors: | Binkowski, T.A, Duggan, E, Shui, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-31 | | Release date: | 2007-09-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Protein of unknown function from Porphyromonas gingivalis W83.
TO BE PUBLISHED
|
|
2QBV
 
 | | Crystal Structure of Intracellular Chorismate Mutase from Mycobacterium Tuberculosis | | Descriptor: | CHORISMATE MUTASE | | Authors: | Ladner, J.E, Reddy, P.T, Kim, S.K, Reddy, S.-K, Nelson, B.C. | | Deposit date: | 2007-06-18 | | Release date: | 2007-06-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A comparative biochemical and structural analysis of the intracellular chorismate mutase (Rv0948c) from Mycobacterium tuberculosis H(37)R(v) and the secreted chorismate mutase (y2828) from Yersinia pestis.
Febs J., 275, 2008
|
|
4LWL
 
 | |
2QDX
 
 | | P.Aeruginosa Fpr with FAD | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin reductase, SULFATE ION | | Authors: | Han, H, Schonbrunn, E. | | Deposit date: | 2007-06-21 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Biochemical and Structural Characterization of Pseudomonas aeruginosa Bfd and FPR: Ferredoxin NADP(+) Reductase and Not Ferredoxin Is the Redox Partner of Heme Oxygenase under Iron-Starvation Conditions
Biochemistry, 46, 2007
|
|
2QIR
 
 | | Crystal structure of aminoglycoside acetyltransferase AAC(6')-Ib in complex whith coenzyme A and kanamycin | | Descriptor: | (1R,2S,3S,4R,6S)-4,6-DIAMINO-3-[(3-AMINO-3-DEOXY-ALPHA-D-GLUCOPYRANOSYL)OXY]-2-HYDROXYCYCLOHEXYL 2,6-DIAMINO-2,6-DIDEOXY-ALPHA-D-GLUCOPYRANOSIDE, Aminoglycoside 6-N-acetyltransferase type Ib11, COENZYME A | | Authors: | Maurice, F, Broutin, I, Podglajen, I, Benas, P, Collatz, E, Dardel, F. | | Deposit date: | 2007-07-05 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Enzyme structural plasticity and the emergence of broad-spectrum antibiotic resistance.
Embo Rep., 9, 2008
|
|
4UZ6
 
 | | STRUCTURE OF THE WNT DEACYLASE NOTUM - CRYSTAL FORM V - SOS COMPLEX - 1.9A | | Descriptor: | 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Zebisch, M, Jones, E.Y. | | Deposit date: | 2014-09-04 | | Release date: | 2015-02-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Notum Deacylates Wnt Proteins to Suppress Signalling Activity.
Nature, 519, 2015
|
|
