7ZTR
 
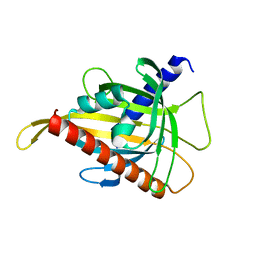 | | Crystal structure of the carotenoid-binding protein domain from silkworm Bombyx mori (BmCBP) in the apoform, W232F mutant | | Descriptor: | Carotenoid-binding protein | | Authors: | Sluchanko, N.N, Boyko, K.M, Varfolomeeva, L.A, Slonimskiy, Y.B, Egorkin, N.A, Maksimov, E.G, Popov, V.O. | | Deposit date: | 2022-05-11 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Silkworm carotenoprotein as an efficient carotenoid extractor, solubilizer and transporter.
Int.J.Biol.Macromol., 223, 2022
|
|
4U2L
 
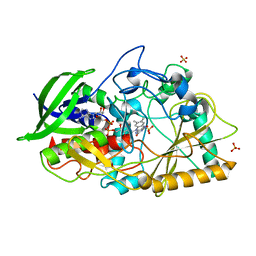 | | Dithionite reduced cholesterol in complex with sulfite | | Descriptor: | (S)-10-((2S,3S,4R)-5-((S)-((S)-(((2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3,4-DIHYDROXY-TETRAHYDROFURAN-2-YL)METHOXY)(HYDROXY)PHOSPHORYLOXY)(HYDROXY)PHOSPHORYLOXY)-2,3,4-TRIHYDROXYPENTYL)-7,8-DIMETHYL-2,4-DIOXO-2,3,4,4A-TETRAHYDROBENZO[G]PTERIDINE-5(10H)-SULFONIC ACID, Cholesterol oxidase, SULFATE ION | | Authors: | Golden, E.A, Vrielink, A. | | Deposit date: | 2014-07-17 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.337 Å) | | Cite: | High-resolution structures of cholesterol oxidase in the reduced state provide insights into redox stabilization.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6BMC
 
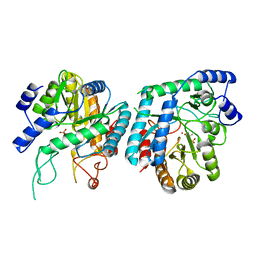 | | The structure of a dimeric type II DAH7PS associated with pyocyanin biosynthesis in Pseudomonas aeruginosa | | Descriptor: | CHLORIDE ION, COBALT (II) ION, PHOSPHOENOLPYRUVATE, ... | | Authors: | Sterritt, O.W, Jameson, G.B, Parker, E.J. | | Deposit date: | 2017-11-14 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and functional characterisation of the entry point to pyocyanin biosynthesis inPseudomonas aeruginosadefines a new 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase subclass.
Biosci. Rep., 38, 2018
|
|
4FUA
 
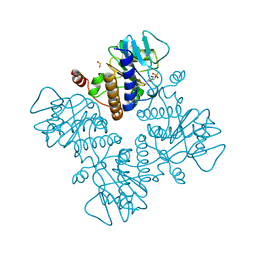 | | L-FUCULOSE-1-PHOSPHATE ALDOLASE COMPLEX WITH PGH | | Descriptor: | BETA-MERCAPTOETHANOL, L-FUCULOSE-1-PHOSPHATE ALDOLASE, PHOSPHOGLYCOLOHYDROXAMIC ACID, ... | | Authors: | Dreyer, M.K, Schulz, G.E. | | Deposit date: | 1996-02-14 | | Release date: | 1996-10-14 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Catalytic mechanism of the metal-dependent fuculose aldolase from Escherichia coli as derived from the structure.
J.Mol.Biol., 259, 1996
|
|
5KXI
 
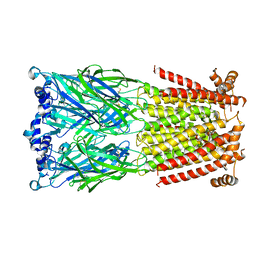 | | X-ray structure of the human Alpha4Beta2 nicotinic receptor | | Descriptor: | (S)-3-(1-METHYLPYRROLIDIN-2-YL)PYRIDINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Neuronal acetylcholine receptor subunit alpha-4, ... | | Authors: | Morales-Perez, C.L, Noviello, C.M, Hibbs, R.E. | | Deposit date: | 2016-07-20 | | Release date: | 2016-09-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.941 Å) | | Cite: | X-ray structure of the human alpha 4 beta 2 nicotinic receptor.
Nature, 538, 2016
|
|
6NJL
 
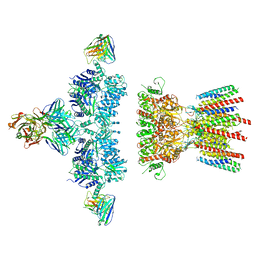 | |
7UTL
 
 | | ALTERNATIVE MODELING OF TROPOMYOSIN IN HUMAN CARDIAC THIN FILAMENT IN THE CALCIUM FREE STATE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Rynkiewicz, M.J, Pavadai, E, Lehman, W. | | Deposit date: | 2022-04-27 | | Release date: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Protein-Protein Docking Reveals Dynamic Interactions of Tropomyosin on Actin Filaments.
Biophys J, 119, 2020
|
|
6WQI
 
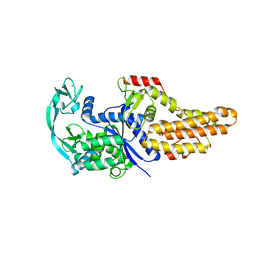 | | Xanthomonas citri Methionyl-tRNA synthetase (apo) | | Descriptor: | Methionine--tRNA ligase, ZINC ION | | Authors: | Mercaldi, G.F, Benedetti, C.E. | | Deposit date: | 2020-04-28 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for diaryldiamine selectivity and competition with tRNA in a type 2 methionyl-tRNA synthetase from a Gram-negative bacterium.
J.Biol.Chem., 296, 2021
|
|
4GOL
 
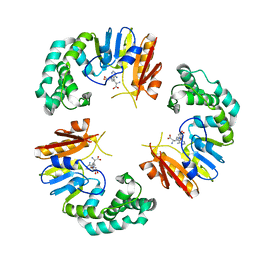 | |
3NP8
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS L36A at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Caro, J.A, Schlessman, J.L, Garcia-Moreno, E.B, Heroux, A. | | Deposit date: | 2010-06-28 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Cavities determine the pressure unfolding of proteins.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2W1I
 
 | | Structure determination of Aurora Kinase in complex with inhibitor | | Descriptor: | 4-[(2-{4-[(CYCLOPROPYLCARBAMOYL)AMINO]-1H-PYRAZOL-3-YL}-1H-BENZIMIDAZOL-6-YL)METHYL]MORPHOLIN-4-IUM, JAK2 | | Authors: | Howard, S, Berdini, V, Boulstridge, J.A, Carr, M.G, Cross, D.M, Curry, J, Devine, L.A, Early, T.R, Fazal, L, Gill, A.L, Heathcote, M, Maman, S, Matthews, J.E, McMenamin, R.L, Navarro, E.F, O'Brien, M.A, O'Reilly, M, Rees, D.C, Reule, M, Tisi, D, Williams, G, Vinkovic, M, Wyatt, P.G. | | Deposit date: | 2008-10-17 | | Release date: | 2009-01-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Fragment-Based Discovery of the Pyrazol-4-Yl Urea (at9283), a Multitargeted Kinase Inhibitor with Potent Aurora Kinase Activity.
J.Med.Chem., 52, 2009
|
|
5KPW
 
 | | Structure of RelA bound to ribosome in presence of A/R tRNA (Structure III) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Bah, E, Madireddy, R, Zhang, Y, Brilot, A.F, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2016-07-05 | | Release date: | 2016-09-28 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Ribosome•RelA structures reveal the mechanism of stringent response activation.
Elife, 5, 2016
|
|
2Y69
 
 | | Bovine heart cytochrome c oxidase re-refined with molecular oxygen | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, CHOLIC ACID, ... | | Authors: | Kaila, V.R.I, Oksanen, E, Goldman, A, Verkhovsky, M.I, Sundholm, D, Wikstrom, M. | | Deposit date: | 2011-01-20 | | Release date: | 2011-02-23 | | Last modified: | 2019-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Combined Quantum Chemical and Crystallographic Study on the Oxidized Binuclear Center of Cytochrome C Oxidase.
Biochim.Biophys.Acta, 1807, 2011
|
|
5TMM
 
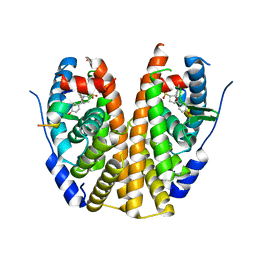 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the OBHS-ASC analog, (E)-6-(4-((1R,4S,6R)-6-((4-bromophenoxy)sulfonyl)-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl)phenyl)hex-5-enoic acid | | Descriptor: | 6-{4-[(1S,4S,6S)-6-[(4-bromophenoxy)sulfonyl]-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl]phenyl}hex-5-enoic acid, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-13 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
7UMV
 
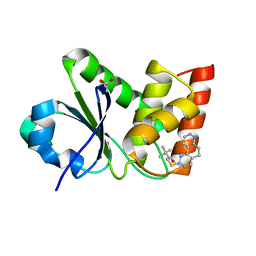 | | Structure of MAP kinase phosphatase 5 in complex with 3,3-dimethyl-1-((5,6-dihydropyrido[2,3-h]quinazolin-2-yl)thio)butan-2-one, an allosteric inhibitor | | Descriptor: | 1-{[(10aP)-5,6-dihydropyrido[2,3-h]quinazolin-2-yl]sulfanyl}-3,3-dimethylbutan-2-one, ACETATE ION, Dual specificity protein phosphatase 10 | | Authors: | Gannam, Z.T.K, Jamali, H, Lolis, E, Anderson, K.S, Ellman, J.A, Bennett, A.M. | | Deposit date: | 2022-04-07 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Defining the structure-activity relationship for a novel class of allosteric MKP5 inhibitors.
Eur.J.Med.Chem., 243, 2022
|
|
4U6Q
 
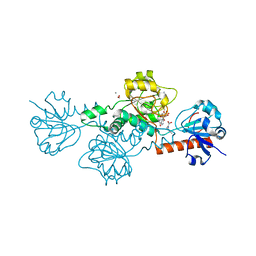 | | CtBP1 bound to inhibitor 2-(hydroxyimino)-3-phenylpropanoic acid | | Descriptor: | (2E)-2-(hydroxyimino)-3-phenylpropanoic acid, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, C-terminal-binding protein 1, ... | | Authors: | Hilbert, B.J, Morris, B.L, Ellis, K.C, Paulsen, J.L, Schiffer, C.A, Grossman, S.R, Royer Jr, W.E. | | Deposit date: | 2014-07-29 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Guided Design of a High Affinity Inhibitor to Human CtBP.
Acs Chem.Biol., 10, 2015
|
|
5A5P
 
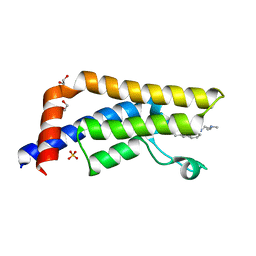 | | Crystal structure of human ATAD2 bromodomain in complex with 8-2-(dimethylamino)ethylamino-3-methyl-1,2-dihydroquinolin-2-one | | Descriptor: | 1,2-ETHANEDIOL, 8-{[2-(dimethylamino)ethyl]amino}-3-methyl-1,2-dihydroquinolin-2-one, ATPASE FAMILY AAA DOMAIN-CONTAINING PROTEIN 2, ... | | Authors: | Chung, C, Bamborough, P, Demont, E. | | Deposit date: | 2015-06-20 | | Release date: | 2015-07-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Fragment-Based Discovery of Low-Micromolar Atad2 Bromodomain Inhibitors.
J.Med.Chem., 58, 2015
|
|
7YVZ
 
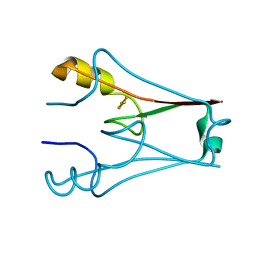 | | Structure of Caenorhabditis elegans CISD-1/mitoNEET | | Descriptor: | CDGSH iron-sulfur domain-containing protein 1 (CISD-1/mitoNEET), FE2/S2 (INORGANIC) CLUSTER | | Authors: | Hasegawa, K, Hagiuda, E, Taguchi, A.T, Geldenhuys, W, Iwasaki, T, Kumasaka, T. | | Deposit date: | 2022-08-20 | | Release date: | 2022-10-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and biological evaluation of Caenorhabditis elegans CISD-1/mitoNEET, a KLP-17 tail domain homologue, supports attenuation of paraquat-induced oxidative stress through a p38 MAPK-mediated antioxidant defense response.
Adv Redox Res, 6, 2022
|
|
4H2O
 
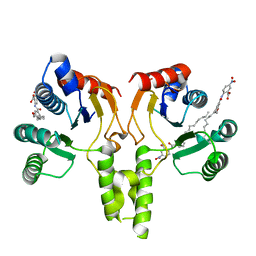 | |
4GON
 
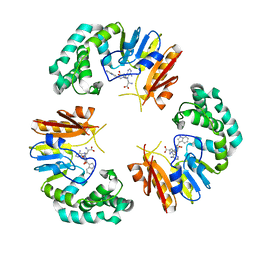 | |
6NQA
 
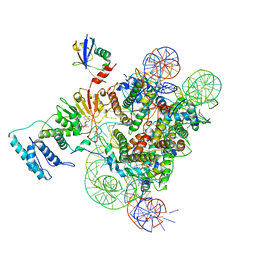 | | Active state Dot1L bound to the H2B-Ubiquitinated nucleosome, 1-to-1 complex | | Descriptor: | 601 DNA Strand 1, 601 DNA Strand 2, Histone H2A type 1, ... | | Authors: | Worden, E.J, Hoffmann, N.A, Wolberger, C. | | Deposit date: | 2019-01-19 | | Release date: | 2019-02-20 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Mechanism of Cross-talk between H2B Ubiquitination and H3 Methylation by Dot1L.
Cell, 176, 2019
|
|
5HQ3
 
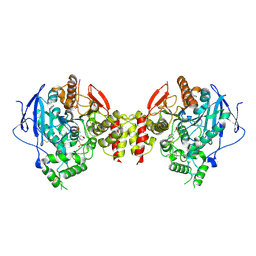 | | Stable, high-expression variant of human acetylcholinesterase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Acetylcholinesterase, O-ETHYLMETHYLPHOSPHONIC ACID ESTER GROUP | | Authors: | Goldenzweig, A, Goldsmith, M, Hill, S.E, Gertman, O, Laurino, P, Ashani, Y, Dym, O, Albeck, S, Unger, T, Prilusky, J, Lieberman, R.L, Aharoni, A, Silman, I, Sussman, J.L, Tawfik, D.S, Fleishman, S.J. | | Deposit date: | 2016-01-21 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Automated Structure- and Sequence-Based Design of Proteins for High Bacterial Expression and Stability.
Mol.Cell, 63, 2016
|
|
7MO7
 
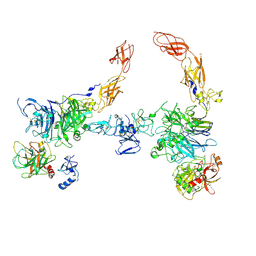 | | Cryo-EM structure of 2:2 c-MET/HGF holo-complex | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, Hepatocyte growth factor, Hepatocyte growth factor receptor | | Authors: | Uchikawa, E, Chen, Z.M, Xiao, G.Y, Zhang, X.W, Bai, X.C. | | Deposit date: | 2021-05-01 | | Release date: | 2021-06-09 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural basis of the activation of c-MET receptor.
Nat Commun, 12, 2021
|
|
4U6T
 
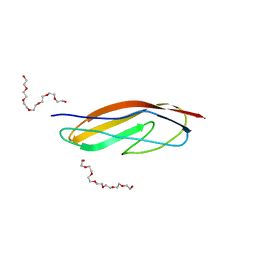 | | Crystal structure of the Clostridium histolyticum colH collagenase polycystic kidney disease-like domain 2a at 1.76 Angstrom resolution | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, ColH protein | | Authors: | Bauer, R, Janowska, K, Sakon, J, Matsushita, O, Latimer, E. | | Deposit date: | 2014-07-29 | | Release date: | 2015-03-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structures of three polycystic kidney disease-like domains from Clostridium histolyticum collagenases ColG and ColH.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5HRY
 
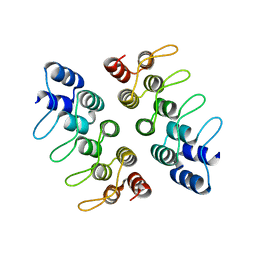 | | Computationally Designed Cyclic Dimer ank3C2_1 | | Descriptor: | ank3C2_1 | | Authors: | Cascio, D, McNamara, D.E, Fallas, J.A, Baker, D, Yeates, T.O. | | Deposit date: | 2016-01-24 | | Release date: | 2017-04-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Computational design of self-assembling cyclic protein homo-oligomers.
Nat Chem, 9, 2017
|
|
