7ZMH
 
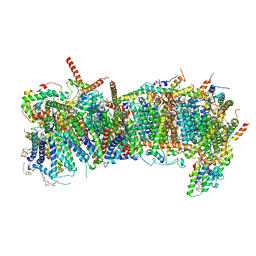 | | CryoEM structure of mitochondrial complex I from Chaetomium thermophilum (state 1) - membrane arm | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Laube, E, Kuehlbrandt, W. | | Deposit date: | 2022-04-19 | | Release date: | 2022-11-30 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (2.47 Å) | | Cite: | Conformational changes in mitochondrial complex I of the thermophilic eukaryote Chaetomium thermophilum.
Sci Adv, 8, 2022
|
|
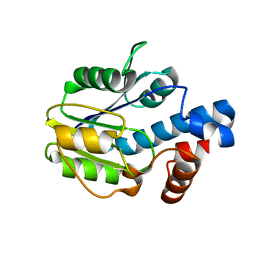
6BWG
 
 | |
5CWV
 
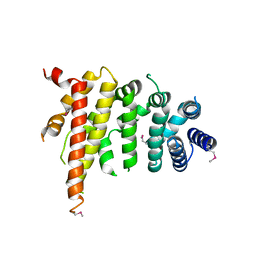 | | Crystal structure of Chaetomium thermophilum Nup192 TAIL domain | | Descriptor: | Nucleoporin NUP192 | | Authors: | Stuwe, T, Bley, C.J, Thierbach, K, Petrovic, S, Schilbach, S, Mayo, D.J, Perriches, T, Rundlet, E.J, Jeon, Y.E, Collins, L.N, Lin, D.H, Paduch, M, Koide, A, Lu, V, Fischer, J, Hurt, E, Koide, S, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2015-07-28 | | Release date: | 2015-09-16 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (3.155 Å) | | Cite: | Architecture of the fungal nuclear pore inner ring complex.
Science, 350, 2015
|
|
8A1F
 
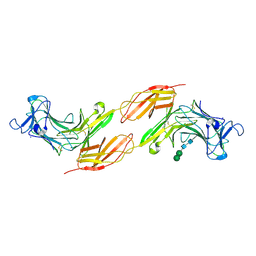 | | Human PTPRK N-terminal domains MAM-Ig-FN1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hay, I.M, Graham, S.C, Sharpe, H.J, Deane, J.E. | | Deposit date: | 2022-06-01 | | Release date: | 2023-03-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Determinants of receptor tyrosine phosphatase homophilic adhesion: Structural comparison of PTPRK and PTPRM extracellular domains.
J.Biol.Chem., 299, 2023
|
|
7KN8
 
 | | Crystal structure of the GH74 xyloglucanase from Xanthomonas campestris (Xcc1752) | | Descriptor: | 1,2-ETHANEDIOL, Cellulase, IODIDE ION, ... | | Authors: | Araujo, E.A, Vieira, P.S, Murakami, M.T, Polikarpov, I. | | Deposit date: | 2020-11-04 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Xyloglucan processing machinery in Xanthomonas pathogens and its role in the transcriptional activation of virulence factors
Nature Communications, 12, 2021
|
|
1E1X
 
 | |
8A17
 
 | | Human PTPRM domains FN3-4, in spacegroup P3221 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Receptor-type tyrosine-protein phosphatase mu, ... | | Authors: | Shamin, M, Graham, S.C, Sharpe, H.J, Deane, J.E. | | Deposit date: | 2022-05-31 | | Release date: | 2023-03-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Determinants of receptor tyrosine phosphatase homophilic adhesion: Structural comparison of PTPRK and PTPRM extracellular domains.
J.Biol.Chem., 299, 2023
|
|
6OGG
 
 | | 70S termination complex with RF2 bound to the UGA codon. Rotated ribosome with RF2 bound (Structure IV). | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Svidritskiy, E, Demo, G, Loveland, A.B, Xu, C, Korostelev, A.A. | | Deposit date: | 2019-04-02 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Extensive ribosome and RF2 rearrangements during translation termination.
Elife, 8, 2019
|
|
6OI3
 
 | | Crystal structure of human WDR5 in complex with monomethyl H3R2 peptide | | Descriptor: | GLYCEROL, Monomethyl H3R2 peptide, SULFATE ION, ... | | Authors: | Lorton, B.M, Harijan, R.K, Burgos, E, Bonanno, J.B, Almo, S.C, Shechter, D. | | Deposit date: | 2019-04-08 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | A Binary Arginine Methylation Switch on Histone H3 Arginine 2 Regulates Its Interaction with WDR5.
Biochemistry, 59, 2020
|
|
1RR9
 
 | | Catalytic domain of E.coli Lon protease | | Descriptor: | ATP-dependent protease La, SULFATE ION | | Authors: | Botos, I, Melnikov, E.E, Cherry, S, Tropea, J.E, Khalatova, A.G, Dauter, Z, Maurizi, M.R, Rotanova, T.V, Wlodawer, A, Gustchina, A. | | Deposit date: | 2003-12-08 | | Release date: | 2003-12-23 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The catalytic domain of Escherichia coli Lon protease has a unique fold and a Ser-Lys dyad in the active site
J.Biol.Chem., 279, 2004
|
|
4ZVR
 
 | |
5VK2
 
 | | Structural basis for antibody-mediated neutralization of Lassa virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hastie, K.M, Zandonatti, M.A, Kleinfelter, L.M, Rowland, M.L, Rowland, M.M, Chandra, K, Branco, L.M, Robinson, J.E, Garry, R.F, Saphire, E.O. | | Deposit date: | 2017-04-20 | | Release date: | 2017-05-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.201 Å) | | Cite: | Structural basis for antibody-mediated neutralization of Lassa virus.
Science, 356, 2017
|
|
4FID
 
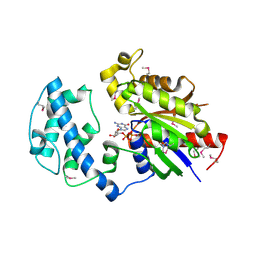 | | Crystal structure of a heterotrimeric G-Protein subunit from entamoeba histolytica, EHG-ALPHA-1 | | Descriptor: | G protein alpha subunit, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Bosch, D.E, Kimple, A.J, Muller, R.E, Gigure, P.M, Willard, F.S, Machius, M, Temple, B.R, Siderovski, D.P. | | Deposit date: | 2012-06-08 | | Release date: | 2012-11-28 | | Last modified: | 2013-06-26 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Heterotrimeric G-protein Signaling Is Critical to Pathogenic Processes in Entamoeba histolytica.
Plos Pathog., 8, 2012
|
|
5WTE
 
 | | Cryo-EM structure for Hepatitis A virus full particle | | Descriptor: | VP1, VP2, VP3 | | Authors: | Wang, X, Zhu, L, Dang, M, Hu, Z, Gao, Q, Yuan, S, Sun, Y, Zhang, B, Ren, J, Walter, T.S, Wang, J, Fry, E.E, Stuart, D.I, Rao, Z. | | Deposit date: | 2016-12-11 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Potent neutralization of hepatitis A virus reveals a receptor mimic mechanism and the receptor recognition site
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6TKX
 
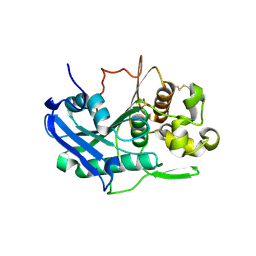 | | Carbohydrate esterase from gut microbiota | | Descriptor: | Carbohydrate esterase, SULFATE ION | | Authors: | Penttinen, L, Hakulinen, N, Master, R.E. | | Deposit date: | 2019-11-29 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Polysaccharide utilization loci-driven enzyme discovery reveals BD-FAE: a bifunctional feruloyl and acetyl xylan esterase active on complex natural xylans.
Biotechnol Biofuels, 14, 2021
|
|
4ZSE
 
 | |
6FZV
 
 | |
2V0D
 
 | | Crystal structure of human CDK2 complexed with a thiazolidinone inhibitor | | Descriptor: | 2-IMINO-5-(1-PYRIDIN-2-YL-METH-(E)-YLIDENE)-1,3-THIAZOLIDIN-4-ONE, CELL DIVISION PROTEIN KINASE 2, CHLORIDE ION | | Authors: | Richardson, C.M, Dokurno, P, Murray, J.B, Surgenor, A.E. | | Deposit date: | 2007-05-14 | | Release date: | 2007-06-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of a Potent Cdk2 Inhibitor with a Novel Binding Mode, Using Virtual Screening and Initial, Structure-Guided Lead Scoping.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
3HK7
 
 | | Crystal structure of uronate isomerase from Bacillus halodurans complexed with zinc and D-Arabinarate, monoclinic crystal form | | Descriptor: | CARBONATE ION, CHLORIDE ION, D-arabinaric acid, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Nguyen, T.T, Raushel, F.M, Almo, S.C. | | Deposit date: | 2009-05-22 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The mechanism of the reaction catalyzed by uronate isomerase illustrates how an isomerase may have evolved from a hydrolase within the amidohydrolase superfamily.
Biochemistry, 48, 2009
|
|
5MLB
 
 | | Crystal structure of human RAS in complex with darpin K27 | | Descriptor: | DARPin K27, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Debreczeni, J.E, Guillard, S, Kolasinska-Zwierz, P, Breed, J, Zhang, J, Bery, N, Marwood, R, Tart, J, Overman, R, Stocki, P, Mistry, B, Phillips, C, Rabbitts, T, Jackson, R, Minter, R. | | Deposit date: | 2016-12-06 | | Release date: | 2017-12-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | INhibition of RAS nucleotide exchange by a DARPin: structural characterisation and effects on downstream signalling by active RAS
To Be Published
|
|
6X87
 
 | | CryoEM structure of the Plasmodium berghei circumsporozoite protein in complex with inhibitory mouse antibody 3D11. | | Descriptor: | 3D11 Fab heavy chain, 3D11 Fab kappa chain, Circumsporozoite protein | | Authors: | Kucharska, I, Thai, E, Rubinstein, J, Julien, J.P. | | Deposit date: | 2020-06-01 | | Release date: | 2020-12-02 | | Last modified: | 2020-12-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural ordering of the Plasmodium berghei circumsporozoite protein repeats by inhibitory antibody 3D11.
Elife, 9, 2020
|
|
2OCS
 
 | | The crystal structure of the first PDZ domain of human NHERF-2 (SLC9A3R2) | | Descriptor: | 1,2-ETHANEDIOL, Na(+)/H(+) exchange regulatory cofactor NHE-RF2, THIOCYANATE ION | | Authors: | Papagrigoriou, E, Phillips, C, Gileadi, C, Elkins, J, Salah, E, Berridge, G, Savitsky, P, Gorrec, F, Umeano, C, Ugochukwu, E, Gileadi, O, Burgess, N, Edwards, A, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Doyle, D.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-12-21 | | Release date: | 2007-01-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of the first PDZ domain of human NHERF-2 (SLC9A3R2)
To be Published
|
|
3NIX
 
 | | Crystal structure of flavoprotein/dehydrogenase from Cytophaga hutchinsonii. Northeast Structural Genomics Consortium Target ChR43. | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Flavoprotein/dehydrogenase | | Authors: | Vorobiev, S, Su, M, Seetharaman, J, Sahdev, S, Xiao, R, Foote, E.L, Ciccosanti, C, Maglaqui, M, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-06-16 | | Release date: | 2010-06-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of flavoprotein/dehydrogenase from Cytophaga hutchinsonii.
To be Published
|
|
7ZW6
 
 | | Oligomeric structure of SynDLP | | Descriptor: | Slr0869 protein | | Authors: | Gewehr, L, Junglas, B, Jilly, R, Franz, J, Wenyu, E.Z, Weidner, T, Bonn, M, Sachse, C, Schneider, D. | | Deposit date: | 2022-05-18 | | Release date: | 2023-04-19 | | Last modified: | 2023-04-26 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | SynDLP is a dynamin-like protein of Synechocystis sp. PCC 6803 with eukaryotic features.
Nat Commun, 14, 2023
|
|
7TBV
 
 | | Crystal structure of the shikimate kinase + 3-dehydroquinate dehydratase + 3-dehydroshikimate dehydrogenase domains of Aro1 from Candida albicans | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Stogios, P.J, Evdokimova, E, Michalska, K, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-12-22 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular analysis and essentiality of Aro1 shikimate biosynthesis multi-enzyme in Candida albicans.
Life Sci Alliance, 5, 2022
|
|
