6XSO
 
 | | GH5-4 broad specificity endoglucanase from an uncultured bacterium | | Descriptor: | (2S)-2-hydroxybutanedioic acid, Cellulase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bingman, C.A, Smith, R.W, Glasgow, E.M, Fox, B.G. | | Deposit date: | 2020-07-15 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A structural and kinetic survey of GH5_4 endoglucanases reveals determinants of broad substrate specificity and opportunities for biomass hydrolysis.
J.Biol.Chem., 295, 2020
|
|
8DD3
 
 | | Human GABAA receptor alpha1-beta2-gamma2 subtype in complex with GABA plus DMCM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GAMMA-AMINO-BUTANOIC ACID, ... | | Authors: | Zhu, S, Hibbs, R.E. | | Deposit date: | 2022-06-17 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural and dynamic mechanisms of GABA A receptor modulators with opposing activities.
Nat Commun, 13, 2022
|
|
8DD2
 
 | | Human GABAA receptor alpha1-beta2-gamma2 subtype in complex with GABA plus Zolpidem | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GAMMA-AMINO-BUTANOIC ACID, ... | | Authors: | Zhu, S, Hibbs, R.E. | | Deposit date: | 2022-06-17 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural and dynamic mechanisms of GABA A receptor modulators with opposing activities.
Nat Commun, 13, 2022
|
|
8FAN
 
 | | Crystal structure of Ky15.1 Fab in complex with circumsporozoite protein KQPA peptide | | Descriptor: | Circumsporozoite protein KQPA peptide, Ky15.1 Antibody, heavy chain, ... | | Authors: | Burn Aschner, C, Thai, E, Julien, J.P. | | Deposit date: | 2022-11-28 | | Release date: | 2023-11-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Molecular determinants of cross-reactivity and potency by VH3-33 antibodies against the Plasmodium falciparum circumsporozoite protein.
Cell Rep, 42, 2023
|
|
6NTA
 
 | | Modified ASL proline bound to Thermus thermophilus 70S (cognate) | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Hoffer, E.D, Maehigashi, T, Subaramanian, S, Hong, S, Dunham, C.M. | | Deposit date: | 2019-01-28 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural insights into mRNA reading frame regulation by tRNA modification and slippery codon-anticodon pairing.
Elife, 9, 2020
|
|
8F9V
 
 | | Crystal structure of Ky15.8 Fab in complex with circumsporozoite protein KQPA peptide | | Descriptor: | 1,2-ETHANEDIOL, Circumsporozoite protein KQPA peptide, Ky15.8 Antibody, ... | | Authors: | Thai, E, Prieto, K, Julien, J.P. | | Deposit date: | 2022-11-24 | | Release date: | 2023-11-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Molecular determinants of cross-reactivity and potency by VH3-33 antibodies against the Plasmodium falciparum circumsporozoite protein.
Cell Rep, 42, 2023
|
|
8F9W
 
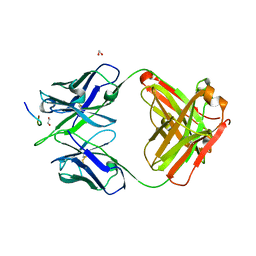 | | Crystal structure of Ky15.8 Fab in complex with circumsporozoite protein NPDP peptide | | Descriptor: | 1,2-ETHANEDIOL, Circumsporozoite protein NPDP peptide, Ky15.8 Antibody, ... | | Authors: | Thai, E, Prieto, K, Julien, J.P. | | Deposit date: | 2022-11-24 | | Release date: | 2023-11-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular determinants of cross-reactivity and potency by VH3-33 antibodies against the Plasmodium falciparum circumsporozoite protein.
Cell Rep, 42, 2023
|
|
6GVF
 
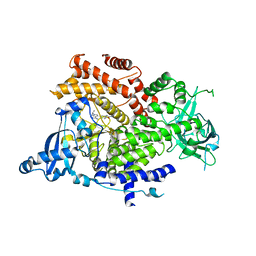 | | Crystal structure of PI3K alpha in complex with 3-(2-Amino-benzooxazol-5-yl)-1-isopropyl-1H-pyrazolo[3,4-d]pyrimidin-4-ylamine | | Descriptor: | 5-(4-azanyl-1-propan-2-yl-pyrazolo[3,4-d]pyrimidin-3-yl)-1,3-benzoxazol-2-amine, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Ouvry, G, Aurelly, M, Bonnary, L, Borde, E, Bouix-Peter, C, Chantalat, L, Clary, L, Defoin-Platel, C, Deret, S, Forissier, M, Harris, C.S, Isabet, T, Lamy, L, Luzy, A.P, Pascau, J, Soulet, C, Taddei, A, Taquet, N, Tomas, L, Thoreau, E, Varvier, E, Vial, E, Hennequin, L.F. | | Deposit date: | 2018-06-21 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Impact of Minor Structural Modifications on Properties of a Series of mTOR Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
6W3A
 
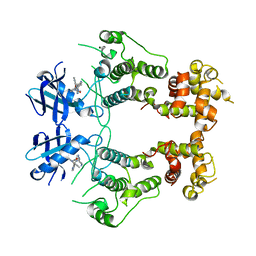 | | Structure of unphosphorylated IRE1 in complex with G-7658 | | Descriptor: | Serine/threonine-protein kinase/endoribonuclease IRE1, ~{N}-[6-methyl-5-[3-[2-[[(3~{S})-piperidin-3-yl]amino]pyrimidin-4-yl]pyridin-2-yl]oxy-naphthalen-1-yl]but-2-ynamide | | Authors: | Ferri, E, Wang, W, Joachim, R, Mortara, K. | | Deposit date: | 2020-03-09 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.606 Å) | | Cite: | Activation of the IRE1 RNase through remodeling of the kinase front pocket by ATP-competitive ligands.
Nat Commun, 11, 2020
|
|
5MDX
 
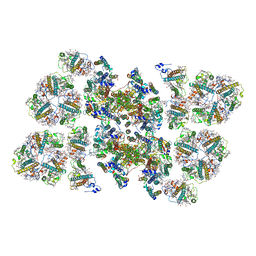 | | Cryo-EM structure of the PSII supercomplex from Arabidopsis thaliana | | Descriptor: | CHLOROPHYLL A, CHLOROPHYLL B, Chlorophyll a-b binding protein 1, ... | | Authors: | van Bezouwen, L.S, Caffarri, S, Kale, R.S, Kouril, R, Thunnissen, A.M.W.H, Oostergetel, G.T, Boekema, E.J. | | Deposit date: | 2016-11-13 | | Release date: | 2017-06-21 | | Last modified: | 2019-04-24 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Subunit and chlorophyll organization of the plant photosystem II supercomplex.
Nat Plants, 3, 2017
|
|
6SSI
 
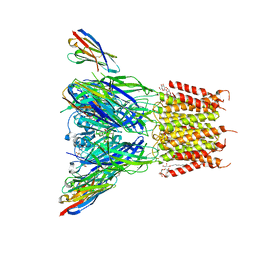 | | Structure of the pentameric ligand-gated ion channel ELIC in complex with a PAM nanobody | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETATE ION, CALCIUM ION, ... | | Authors: | Ulens, C, Brams, M, Evans, G.L, Spurny, R, Govaerts, C, Pardon, E, Steyaert, J. | | Deposit date: | 2019-09-07 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Modulation of the Erwinia ligand-gated ion channel (ELIC) and the 5-HT 3 receptor via a common vestibule site.
Elife, 9, 2020
|
|
5JEA
 
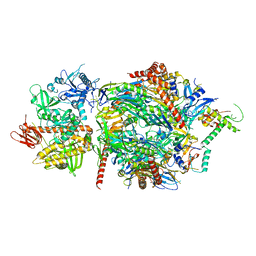 | | Structure of a cytoplasmic 11-subunit RNA exosome complex including Ski7, bound to RNA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Exosome complex component CSL4, Exosome complex component MTR3, ... | | Authors: | Kowalinski, E, Ebert, J, Stegmann, E, Conti, E. | | Deposit date: | 2016-04-18 | | Release date: | 2016-07-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of a Cytoplasmic 11-Subunit RNA Exosome Complex.
Mol.Cell, 63, 2016
|
|
5J3C
 
 | |
6VZQ
 
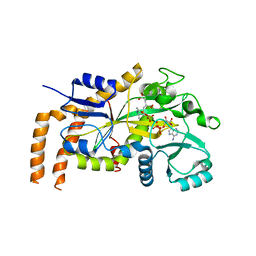 | | Engineered TTLL6 mutant bound to alpha-elongation analog | | Descriptor: | (2~{S})-2-[[[(1~{R})-1-acetamido-4-oxidanyl-4-oxidanylidene-butyl]-phosphonooxy-phosphoryl]methyl]pentanedioic acid, (2~{S})-2-[[[(1~{S})-1-acetamidoethyl]-phosphonooxy-phosphoryl]methyl]pentanedioic acid, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Mahalingan, K.K, Keenen, E.K, Strickland, E.K, Li, Y, Liu, Y, Ball, H.L, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2020-02-28 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Structural basis for polyglutamate chain initiation and elongation by TTLL family enzymes.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6W23
 
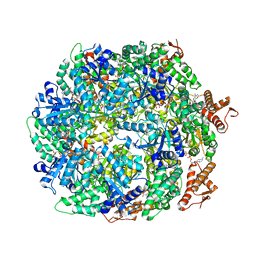 | | ClpA Disengaged State bound to RepA-GFP (Focused Classification) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ... | | Authors: | Lopez, K.L, Rizo, A.N, Tse, E, Lin, J, Scull, N.W, Thwin, A.C, Lucius, A.L, Shorter, J, Southworth, D.R. | | Deposit date: | 2020-03-04 | | Release date: | 2020-05-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Conformational plasticity of the ClpAP AAA+ protease couples protein unfolding and proteolysis.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6FRM
 
 | | Crystal Structure of coenzyme F420H2 oxidase (FprA) co-crystallized with 10 mM Tb-Xo4 | | Descriptor: | CHLORIDE ION, Coenzyme F420H2 oxidase (FprA), FE (III) ION, ... | | Authors: | Engilberge, S, Riobe, F, Wagner, T, Di Pietro, S, Shima, S, Girard, E, Dumont, E, Maury, O. | | Deposit date: | 2018-02-16 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Unveiling the Binding Modes of the Crystallophore, a Terbium-based Nucleating and Phasing Molecular Agent for Protein Crystallography.
Chemistry, 24, 2018
|
|
8A9Z
 
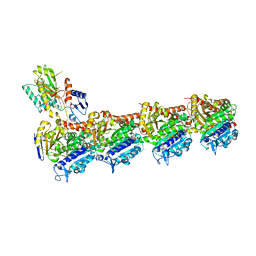 | | Tubulin-[1,2]oxazoloisoindole-2e complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 7-[(3,5-dimethoxyphenyl)methyl]pyrrolo[3,4-g][1,2]benzoxazole, CALCIUM ION, ... | | Authors: | Prota, A.E, Abel, A.-C, Steinmetz, M.O, Barraja, P, Montalbano, A, Spano, V. | | Deposit date: | 2022-06-29 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.294 Å) | | Cite: | Development of [1,2]oxazoloisoindoles tubulin polymerization inhibitors: Further chemical modifications and potential therapeutic effects against lymphomas.
Eur.J.Med.Chem., 243, 2022
|
|
6W3C
 
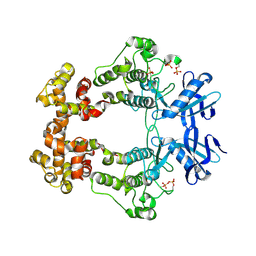 | | Structure of phosphorylated apo IRE1 | | Descriptor: | Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Wallweber, H, Mortara, K, Ferri, E, Wang, W, Rudolph, J. | | Deposit date: | 2020-03-09 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Activation of the IRE1 RNase through remodeling of the kinase front pocket by ATP-competitive ligands.
Nat Commun, 11, 2020
|
|
6NSZ
 
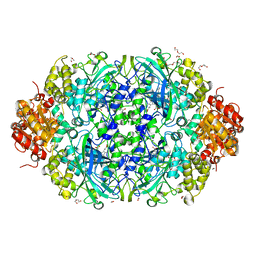 | | X-ray reduced Catalase 3 from N.Crassa (0.526 MGy) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Catalase-3, ... | | Authors: | Zarate-Romero, A, Rudino-Pinera, E, Stojanoff, V. | | Deposit date: | 2019-01-27 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray driven reduction of Cpd I of Catalase-3 from N. crassa reveals differential sensitivity of active sites and formation of ferrous state.
Arch.Biochem.Biophys., 666, 2019
|
|
8FF3
 
 | |
7TIY
 
 | | Crystal structure of SARS-CoV-2 3CL in complex with inhibitor NK01-48 | | Descriptor: | (1S,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-[(N-{[(2,4,5-trifluorophenyl)methoxy]carbonyl}-L-leucyl)amino]propane-1-sulfonic acid, 1,2-ETHANEDIOL, 3C-like proteinase nsp5, ... | | Authors: | Forouhar, F, Liu, H, Iketani, S, Zack, A, Khanizeman, N, Bednarova, E, Fowler, B, Hong, S.J, Mohri, H, Nair, M.S, Huang, Y, Tay, N.E.S, Lee, S, Karan, C, Resnick, S.J, Quinn, C, Li, W, Shion, H, Jurtschenko, C, Lauber, M.A, McDonald, T, Stokes, M.E, Hurst, B, Rovis, T, Chavez, A, Ho, D.D, Stockwell, B.R. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Development of optimized drug-like small molecule inhibitors of the SARS-CoV-2 3CL protease for treatment of COVID-19.
Nat Commun, 13, 2022
|
|
6VUI
 
 | | Metabolite-bound PreQ1 riboswitch with Mn2+ | | Descriptor: | 7-DEAZA-7-AMINOMETHYL-GUANINE, MANGANESE (II) ION, PREQ1 RIBOSWITCH | | Authors: | Jenkins, J.L, Wedekind, J.E. | | Deposit date: | 2020-02-15 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.681 Å) | | Cite: | Analysis of a preQ1-I riboswitch in effector-free and bound states reveals a metabolite-programmed nucleobase-stacking spine that controls gene regulation.
Nucleic Acids Res., 48, 2020
|
|
6NOJ
 
 | | PD-L1 IgV domain V76T with fragment | | Descriptor: | Programmed cell death 1 ligand 1, methyl 3-amino-4-(2-fluorophenyl)-1H-pyrrole-2-carboxylate | | Authors: | Zhao, B, Perry, E. | | Deposit date: | 2019-01-16 | | Release date: | 2019-02-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Fragment-based screening of programmed death ligand 1 (PD-L1).
Bioorg. Med. Chem. Lett., 29, 2019
|
|
7TIU
 
 | | Crystal structure of SARS-CoV-2 3CL in complex with inhibitor EB46 | | Descriptor: | (1S,2S)-2-[(N-{[(3-chlorophenyl)methoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5, MAGNESIUM ION, ... | | Authors: | Forouhar, F, Liu, H, Iketani, S, Zack, A, Khanizeman, N, Bednarova, E, Fowler, B, Hong, S.J, Mohri, H, Nair, M.S, Huang, Y, Tay, N.E.S, Lee, S, Karan, C, Resnick, S.J, Quinn, C, Li, W, Shion, H, Jurtschenko, C, Lauber, M.A, McDonald, T, Stokes, M.E, Hurst, B, Rovis, T, Chavez, A, Ho, D.D, Stockwell, B.R. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Development of optimized drug-like small molecule inhibitors of the SARS-CoV-2 3CL protease for treatment of COVID-19.
Nat Commun, 13, 2022
|
|
7TIZ
 
 | | Crystal structure of SARS-CoV-2 3CL in complex with inhibitor NK01-63 | | Descriptor: | (1S,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-{[N-({[3-(trifluoromethyl)phenyl]methoxy}carbonyl)-L-leucyl]amino}propane-1-sulfonic acid, 1,2-ETHANEDIOL, 3C-like proteinase nsp5, ... | | Authors: | Forouhar, F, Liu, H, Iketani, S, Zack, A, Khanizeman, N, Bednarova, E, Fowler, B, Hong, S.J, Mohri, H, Nair, M.S, Huang, Y, Tay, N.E.S, Lee, S, Karan, C, Resnick, S.J, Quinn, C, Li, W, Shion, H, Jurtschenko, C, Lauber, M.A, McDonald, T, Stokes, M.E, Hurst, B, Rovis, T, Chavez, A, Ho, D.D, Stockwell, B.R. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Development of optimized drug-like small molecule inhibitors of the SARS-CoV-2 3CL protease for treatment of COVID-19.
Nat Commun, 13, 2022
|
|
