6RTC
 
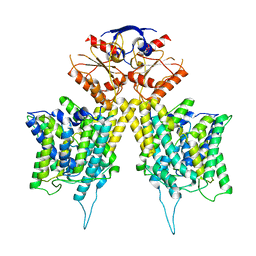 | |
6YTO
 
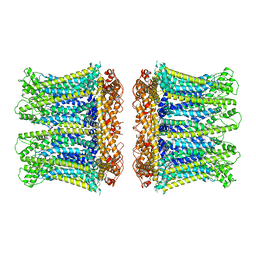 | |
6YTQ
 
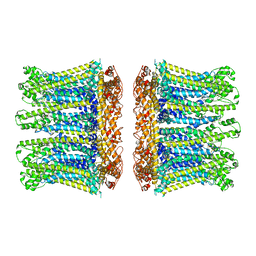 | |
6YTK
 
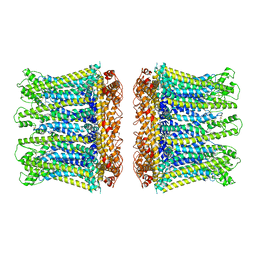 | |
6YTL
 
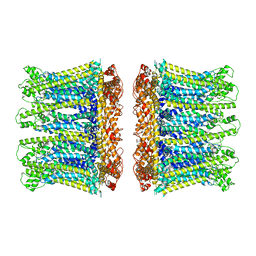 | |
8BEN
 
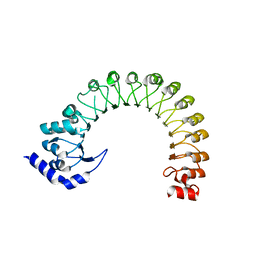 | | LRR domain Structure of the LRRC8C protein | | Descriptor: | Volume-regulated anion channel subunit LRRC8C | | Authors: | Sawicka, M, Dutzler, R. | | Deposit date: | 2022-10-21 | | Release date: | 2022-12-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of a volume-regulated heteromeric LRRC8A/C channel.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8B8K
 
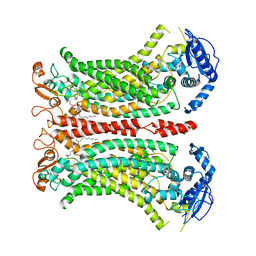 | | Cryo-EM structure of Ca2+-bound mTMEM16F N562A mutant in Digitonin closed/closed | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Anoctamin-6, CALCIUM ION | | Authors: | Arndt, M, Alvadia, C, Straub, M.S, Clerico-Mosina, V, Paulino, C, Dutzler, R. | | Deposit date: | 2022-10-04 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural basis for the activation of the lipid scramblase TMEM16F.
Nat Commun, 13, 2022
|
|
8B8G
 
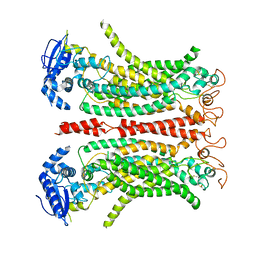 | | Cryo-EM structure of Ca2+-free mTMEM16F F518H mutant in Digitonin | | Descriptor: | Anoctamin-6 | | Authors: | Arndt, M, Alvadia, C, Straub, M.S, Clerico-Mosina, V, Paulino, C, Dutzler, R. | | Deposit date: | 2022-10-04 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structural basis for the activation of the lipid scramblase TMEM16F.
Nat Commun, 13, 2022
|
|
8B8J
 
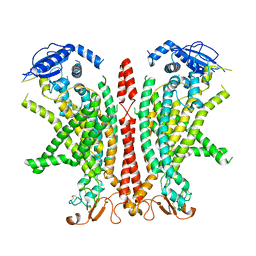 | | Cryo-EM structure of Ca2+-bound mTMEM16F F518H mutant in Digitonin | | Descriptor: | Anoctamin-6, CALCIUM ION | | Authors: | Arndt, M, Alvadia, C, Straub, M.S, Clerico Mosina, V, Paulino, C, Dutzler, R. | | Deposit date: | 2022-10-04 | | Release date: | 2022-11-16 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Structural basis for the activation of the lipid scramblase TMEM16F.
Nat Commun, 13, 2022
|
|
8BC0
 
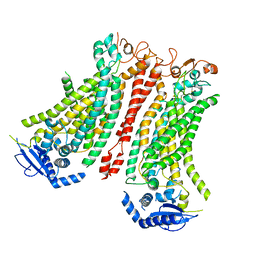 | | Cryo-EM structure of Ca2+-bound mTMEM16F F518A Q623A mutant in GDN open/closed | | Descriptor: | Anoctamin-6, CALCIUM ION | | Authors: | Arndt, M, Alvadia, C, Straub, M, Clerico-Mosina, V, Paulino, C, Dutzler, R. | | Deposit date: | 2022-10-14 | | Release date: | 2022-11-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural basis for the activation of the lipid scramblase TMEM16F.
Nat Commun, 13, 2022
|
|
8BC1
 
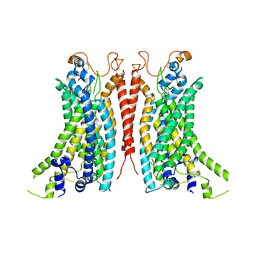 | | Cryo-EM Structure of Ca2+-bound mTMEM16F F518A_Q623A mutant in GDN | | Descriptor: | Anoctamin-6,mTMEM16F, CALCIUM ION | | Authors: | Arndt, M, Alvadia, C, Straub, M.S, Clerico-Mosina, V, Paulino, C, Dutzler, R. | | Deposit date: | 2022-10-14 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural basis for the activation of the lipid scramblase TMEM16F.
Nat Commun, 13, 2022
|
|
8B8M
 
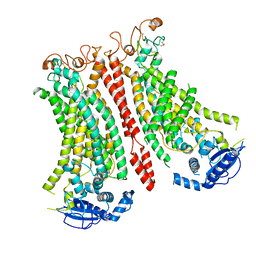 | | Cryo-EM structure of Ca2+-bound mTMEM16F N562A mutant in Digitonin open/closed | | Descriptor: | Anoctamin-6, CALCIUM ION | | Authors: | Arndt, M, Alvadia, C, Straub, M.S, Clerico-Mosina, V, Paulino, C, Dutzler, R. | | Deposit date: | 2022-10-04 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structural basis for the activation of the lipid scramblase TMEM16F.
Nat Commun, 13, 2022
|
|
8B40
 
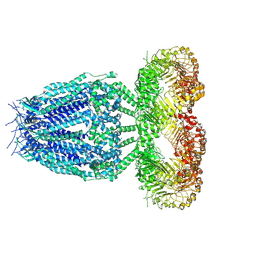 | |
8B41
 
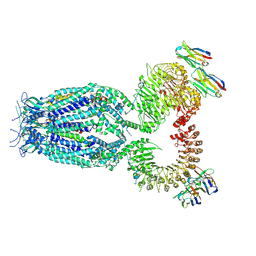 | |
8B8Q
 
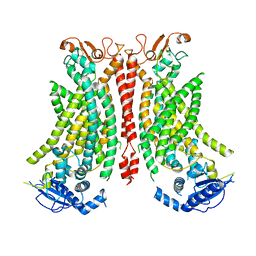 | | Structure of mTMEM16F in lipid Nanodiscs in the presence of Ca2+ | | Descriptor: | Anoctamin-6, CALCIUM ION | | Authors: | Arndt, M, Alvadia, C, Straub, M.S, Clerico-Mosina, V, Paulino, C, Dutzler, R. | | Deposit date: | 2022-10-04 | | Release date: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Structural basis for the activation of the lipid scramblase TMEM16F.
Nat Commun, 13, 2022
|
|
8B42
 
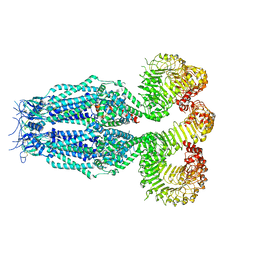 | |
2XQ4
 
 | | Pentameric ligand gated ion channel GLIC in complex with tetramethylarsonium (TMAs) | | Descriptor: | ARSENIC, GLR4197 PROTEIN | | Authors: | Hilf, R.J.C, Bertozzi, C, Zimmermann, I, Reiter, A, Trauner, D, Dutzler, R. | | Deposit date: | 2010-09-01 | | Release date: | 2010-11-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural Basis of Open Channel Block in a Prokaryotic Pentameric Ligand-Gated Ion Channel
Nat.Struct.Mol.Biol., 17, 2010
|
|
8C03
 
 | | Structure of SLC40/ferroportin in complex with vamifeport and synthetic nanobody Sy12 in outward-facing conformation | | Descriptor: | 2-[2-[2-(1~{H}-benzimidazol-2-yl)ethylamino]ethyl]-~{N}-[(3-fluoranylpyridin-2-yl)methyl]-1,3-oxazole-4-carboxamide, Solute carrier family 40 member 1 | | Authors: | Lehmann, E.F, Liziczai, M, Drozdzyk, K, Dutzler, R, Manatschal, C. | | Deposit date: | 2022-12-15 | | Release date: | 2023-03-22 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.89 Å) | | Cite: | Structures of ferroportin in complex with its specific inhibitor vamifeport.
Elife, 12, 2023
|
|
2XQ6
 
 | | Pentameric ligand gated ion channel GLIC in complex with cesium ion (Cs+) | | Descriptor: | CESIUM ION, GLR4197 PROTEIN | | Authors: | Hilf, R.J.C, Bertozzi, C, Zimmermann, I, Reiter, A, Trauner, D, Dutzler, R. | | Deposit date: | 2010-09-01 | | Release date: | 2010-11-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural Basis of Open Channel Block in a Prokaryotic Pentameric Ligand-Gated Ion Channel
Nat.Struct.Mol.Biol., 17, 2010
|
|
2XQA
 
 | | Pentameric ligand gated ion channel GLIC in complex with tetrabutylantimony (TBSb) | | Descriptor: | ANTIMONY (III) ION, GLR4197 PROTEIN | | Authors: | Hilf, R.J.C, Bertozzi, C, Zimmermann, I, Reiter, A, Trauner, D, Dutzler, R. | | Deposit date: | 2010-09-01 | | Release date: | 2010-11-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural Basis of Open Channel Block in a Prokaryotic Pentameric Ligand-Gated Ion Channel
Nat.Struct.Mol.Biol., 17, 2010
|
|
8BZY
 
 | | Structure of SLC40/ferroportin in complex with vamifeport and synthetic nanobody Sy3 in occluded conformation | | Descriptor: | 2-[2-[2-(1~{H}-benzimidazol-2-yl)ethylamino]ethyl]-~{N}-[(3-fluoranylpyridin-2-yl)methyl]-1,3-oxazole-4-carboxamide, DIUNDECYL PHOSPHATIDYL CHOLINE, Solute carrier family 40 member 1, ... | | Authors: | Lehmann, E.F, Liziczai, M, Drozdzyk, K, Dutzler, R, Manatschal, C. | | Deposit date: | 2022-12-15 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Structures of ferroportin in complex with its specific inhibitor vamifeport.
Elife, 12, 2023
|
|
8C02
 
 | | Structure of SLC40/ferroportin in complex with synthetic nanobody Sy3 in occluded conformation | | Descriptor: | Solute carrier family 40 member 1, Sybody3 | | Authors: | Lehmann, E.F, Liziczai, M, Drozdzyk, K, Dutzler, R, Manatschal, C. | | Deposit date: | 2022-12-15 | | Release date: | 2023-03-22 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (4.09 Å) | | Cite: | Structures of ferroportin in complex with its specific inhibitor vamifeport.
Elife, 12, 2023
|
|
2XQ9
 
 | | Pentameric ligand gated ion channel GLIC mutant E221A in complex with tetraethylarsonium (TEAs) | | Descriptor: | ARSENIC, GLR4197 PROTEIN | | Authors: | Hilf, R.J.C, Bertozzi, C, Zimmermann, I, Reiter, A, Trauner, D, Dutzler, R. | | Deposit date: | 2010-09-01 | | Release date: | 2010-11-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis of Open Channel Block in a Prokaryotic Pentameric Ligand-Gated Ion Channel
Nat.Struct.Mol.Biol., 17, 2010
|
|
2XQ8
 
 | | Pentameric ligand gated ion channel GLIC in complex with zinc ion (Zn2+) | | Descriptor: | GLR4197 PROTEIN, ZINC ION | | Authors: | Hilf, R.J.C, Bertozzi, C, Zimmermann, I, Reiter, A, Trauner, D, Dutzler, R. | | Deposit date: | 2010-09-01 | | Release date: | 2010-11-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural Basis of Open Channel Block in a Prokaryotic Pentameric Ligand-Gated Ion Channel
Nat.Struct.Mol.Biol., 17, 2010
|
|
2XQ7
 
 | | Pentameric ligand gated ion channel GLIC in complex with cadmium ion (Cd2+) | | Descriptor: | CADMIUM ION, GLR4197 PROTEIN | | Authors: | Hilf, R.J.C, Bertozzi, C, Zimmermann, I, Reiter, A, Trauner, D, Dutzler, R. | | Deposit date: | 2010-09-01 | | Release date: | 2010-11-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural Basis of Open Channel Block in a Prokaryotic Pentameric Ligand-Gated Ion Channel
Nat.Struct.Mol.Biol., 17, 2010
|
|
