7Y01
 
 | |
5ZWZ
 
 | |
5ZWX
 
 | |
1G2I
 
 | | CRYSTAL STRUCTURE OF A NOVEL INTRACELLULAR PROTEASE FROM PYROCOCCUS HORIKOSHII AT 2 A RESOLUTION | | Descriptor: | PROTEASE I, SULFATE ION | | Authors: | Du, X, Choi, I.-G, Kim, R, Jancarik, J, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2000-10-19 | | Release date: | 2000-11-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of an intracellular protease from Pyrococcus horikoshii at 2-A resolution.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
7YHQ
 
 | |
7YHO
 
 | |
7YHP
 
 | |
1ILW
 
 | |
1IM5
 
 | |
6X28
 
 | | Crystal structure of PT2243 bound to HIF2a-B*:ARNT-B* complex | | Descriptor: | (1R)-4-(3,5-difluorophenoxy)-7-(trifluoromethyl)-2,3-dihydro-1H-inden-1-ol, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Du, X. | | Deposit date: | 2020-05-20 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of PT2243 bound to HIF2-PasB*:ARNT-PasB* complex
To Be Published
|
|
6X37
 
 | | Crystal structure of PT3245 bound to HIF2a-B*:ARNT-B* complex | | Descriptor: | 3-fluoro-5-{[(7R)-7-hydroxy-1-(trifluoromethyl)-6,7-dihydro-5H-cyclopenta[c]pyridin-4-yl]oxy}benzonitrile, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Du, X. | | Deposit date: | 2020-05-21 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of PT3245 bound to HIF2a-B*:ARNT-B* complex
To Be Published
|
|
6X21
 
 | | Crystal structure of PT1673 bound to HIF2a-B*:ARNT-B* complex | | Descriptor: | 1-(3-bromo-5-fluorophenoxy)-4-[(difluoromethyl)sulfonyl]-2-nitrobenzene, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Du, X. | | Deposit date: | 2020-05-19 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystal structure of PT1673 bound to HIF2a-B*:ARNT-B* complex
To Be Published
|
|
6X2H
 
 | | Crystal structure of PT2863 bound to HIF2a-B*:ARNT-B* complex | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1, cis-3-({(1S)-7-[dihydroxy(trifluoromethyl)-lambda~4~-sulfanyl]-2,2-difluoro-1-hydroxy-2,3-dihydro-1H-inden-4-yl}oxy)cyclobutane-1-carbonitrile | | Authors: | Du, X. | | Deposit date: | 2020-05-20 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of PT2863 bound to HIF2a-B*:ARNT-B* complex
To Be Published
|
|
6X3D
 
 | | Crystal structure of PT3388 bound to HIF2a-B*:ARNT-B* complex | | Descriptor: | (6R,7S)-4-[(3,3-difluorocyclobutyl)oxy]-6-fluoro-1-(trifluoromethyl)-6,7-dihydro-5H-cyclopenta[c]pyridin-7-ol, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Du, X. | | Deposit date: | 2020-05-21 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of PT3388 bound to HIF2a-B*:ARNT-B* complex
To Be Published
|
|
8HIL
 
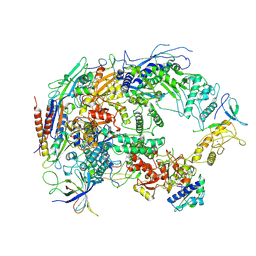 | | A cryo-EM structure of B. oleracea RNA polymerase V at 3.57 Angstrom | | Descriptor: | DNA-dependent RNA polymerase IV and V subunit 2, DNA-directed RNA polymerase V largest subunit, DNA-directed RNA polymerase subunit, ... | | Authors: | Du, X, Xie, G, Hu, H, Du, J. | | Deposit date: | 2022-11-20 | | Release date: | 2023-03-22 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Structure and mechanism of the plant RNA polymerase V.
Science, 379, 2023
|
|
5UFP
 
 | | Crystal structure of PT2399 bound to HIF2a-B*:ARNT-B* complex | | Descriptor: | 3-({(1S)-7-[(difluoromethyl)sulfonyl]-2,2-difluoro-1-hydroxy-2,3-dihydro-1H-inden-4-yl}oxy)-5-fluorobenzonitrile, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Du, X. | | Deposit date: | 2017-01-05 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | On-target efficacy of a HIF-2 alpha antagonist in preclinical kidney cancer models.
Nature, 539, 2016
|
|
5TBM
 
 | | Crystal structure of PT2385 bound to HIF2a-B*:ARNT-B* complex | | Descriptor: | 3-{[(1S)-2,2-difluoro-1-hydroxy-7-(methylsulfonyl)-2,3-dihydro-1H-inden-4-yl]oxy}-5-fluorobenzonitrile, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Du, X. | | Deposit date: | 2016-09-12 | | Release date: | 2016-09-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Small-Molecule Antagonist of HIF2 alpha Is Efficacious in Preclinical Models of Renal Cell Carcinoma.
Cancer Res., 76, 2016
|
|
6CZW
 
 | | Crystal structure of PT1940 bound to HIF2a-B*:ARNT-B* complex | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1, {2-bromo-3-(3-chloro-5-fluorophenoxy)-6-[(difluoromethyl)sulfonyl]phenyl}methanol | | Authors: | Du, X. | | Deposit date: | 2018-04-09 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of PT1940 bound to HIF2a-B*:ARNT-B* complex
To Be Published
|
|
6D09
 
 | | Crystal structure of PT2440 bound to HIF2a-B*:ARNT-B* complex | | Descriptor: | 3-{[(3R)-4-(difluoromethyl)-2,2-difluoro-3-hydroxy-1,1-dioxo-2,3-dihydro-1H-1-benzothiophen-5-yl]oxy}-5-fluorobenzonitrile, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Du, X. | | Deposit date: | 2018-04-10 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of PT1940 bound to HIF2a-B*:ARNT-B* complex
To Be Published
|
|
6D0B
 
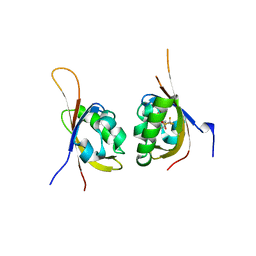 | | Crystal structure of PT1614 bound to HIF2a-B*:ARNT-B* complex | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1, N-(3-chloro-5-fluorophenyl)-2-nitro-4-[(trifluoromethyl)sulfonyl]aniline | | Authors: | Du, X. | | Deposit date: | 2018-04-10 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of PT1614 bound to HIF2a-B*:ARNT-B* complex
To Be Published
|
|
7W82
 
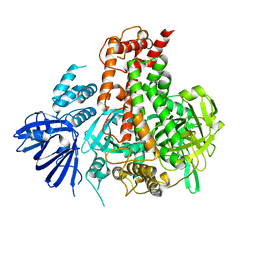 | | Crystal structure of maize RDR2 | | Descriptor: | RNA-dependent RNA polymerase | | Authors: | Du, X, Yang, Z, Du, J. | | Deposit date: | 2021-12-07 | | Release date: | 2022-06-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of plant RNA-DEPENDENT RNA POLYMERASE 2, an enzyme involved in small interfering RNA production.
Plant Cell, 34, 2022
|
|
7W88
 
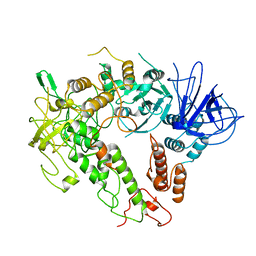 | |
7W84
 
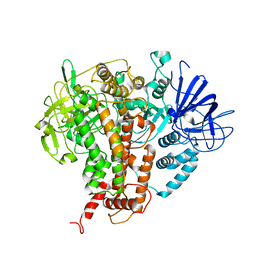 | |
6D0C
 
 | | Crystal structure of HIF2a-B*:ARNT-B* complex | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Du, X. | | Deposit date: | 2018-04-10 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of PT1940 bound to HIF2a-B*:ARNT-B* complex
To Be Published
|
|
7D8U
 
 | | Crystal structure of the C-terminal domain of pNP868R from African swine fever virus | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, GTP--RNA guanylyltransferase, ... | | Authors: | Du, X, Geng, Z, Zhang, H. | | Deposit date: | 2020-10-10 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and Biochemical Characteristic of the Methyltransferase (MTase) Domain of RNA Capping Enzyme from African Swine Fever Virus.
J.Virol., 2020
|
|
