1QLK
 
 | | SOLUTION STRUCTURE OF CA(2+)-LOADED RAT S100B (BETABETA) NMR, 20 STRUCTURES | | 分子名称: | CALCIUM ION, S-100 PROTEIN | | 著者 | Drohat, A.C, Baldisseri, D.M, Rustandi, R.R, Weber, D.J. | | 登録日 | 1997-09-26 | | 公開日 | 1998-11-11 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of calcium-bound rat S100B(betabeta) as determined by nuclear magnetic resonance spectroscopy,.
Biochemistry, 37, 1998
|
|
1SYM
 
 | |
1LMZ
 
 | |
1EUG
 
 | | CRYSTAL STRUCTURE OF ESCHERICHIA COLI URACIL DNA GLYCOSYLASE AND ITS COMPLEXES WITH URACIL AND GLYCEROL: STRUCTURE AND GLYCOSYLASE MECHANISM REVISITED | | 分子名称: | PROTEIN (GLYCOSYLASE) | | 著者 | Xiao, G, Tordova, M, Jagadeesh, J, Drohat, A.C, Stivers, J.T, Gilliland, G.L. | | 登録日 | 1998-10-12 | | 公開日 | 1999-10-12 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal structure of Escherichia coli uracil DNA glycosylase and its complexes with uracil and glycerol: structure and glycosylase mechanism revisited.
Proteins, 35, 1999
|
|
2RBA
 
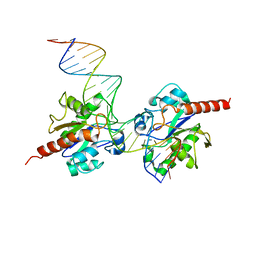 | | Structure of Human Thymine DNA Glycosylase Bound to Abasic and Undamaged DNA | | 分子名称: | DNA (5'-D(*DCP*DAP*DGP*DCP*DTP*DCP*DTP*DGP*DTP*DAP*DCP*DGP*DTP*DGP*DAP*DGP*DCP*DAP*DGP*DTP*DGP*DGP*DA)-3'), DNA (5'-D(*DCP*DCP*DAP*DCP*DTP*DGP*DCP*DTP*DCP*DAP*(3DR)P*DGP*DTP*DAP*DCP*DAP*DGP*DAP*DGP*DCP*DTP*DGP*DT)-3'), G/T mismatch-specific thymine DNA glycosylase | | 著者 | Maiti, A, Pozharski, E, Drohat, A.C. | | 登録日 | 2007-09-18 | | 公開日 | 2008-06-17 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.79 Å) | | 主引用文献 | Crystal structure of human thymine DNA glycosylase bound to DNA elucidates sequence-specific mismatch recognition.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
5T2W
 
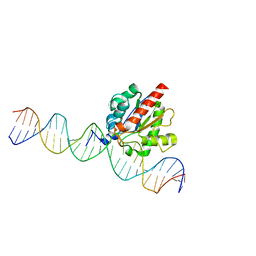 | |
2EUG
 
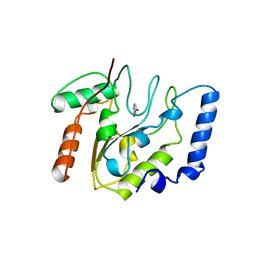 | | CRYSTAL STRUCTURE OF ESCHERICHIA COLI URACIL DNA GLYCOSYLASE AND ITS COMPLEXES WITH URACIL AND GLYCEROL: STRUCTURE AND GLYCOSYLASE MECHANISM REVISITED | | 分子名称: | PROTEIN (GLYCOSYLASE), URACIL | | 著者 | Xiao, G, Tordova, M, Jagadeesh, J, Drohat, A.C, Stivers, J.T, Gilliland, G.L. | | 登録日 | 1998-10-13 | | 公開日 | 1999-10-13 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal structure of Escherichia coli uracil DNA glycosylase and its complexes with uracil and glycerol: structure and glycosylase mechanism revisited.
Proteins, 35, 1999
|
|
5CYS
 
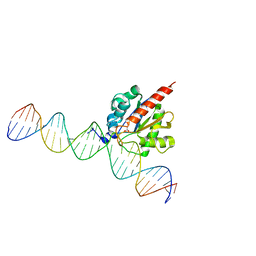 | |
1B4C
 
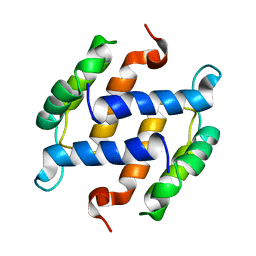 | | SOLUTION STRUCTURE OF RAT APO-S100B USING DIPOLAR COUPLINGS | | 分子名称: | PROTEIN (S-100 PROTEIN, BETA CHAIN) | | 著者 | Weber, D.J, Drohat, A.C, Tjandra, N, Baldisseri, D.M. | | 登録日 | 1998-12-17 | | 公開日 | 1998-12-30 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The use of dipolar couplings for determining the solution structure of rat apo-S100B(betabeta).
Protein Sci., 8, 1999
|
|
1P7M
 
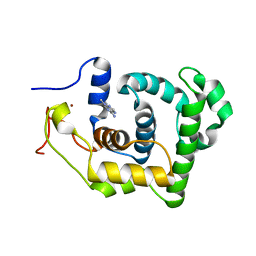 | | SOLUTION STRUCTURE AND BASE PERTURBATION STUDIES REVEAL A NOVEL MODE OF ALKYLATED BASE RECOGNITION BY 3-METHYLADENINE DNA GLYCOSYLASE I | | 分子名称: | 3-METHYL-3H-PURIN-6-YLAMINE, DNA-3-methyladenine glycosylase I, ZINC ION | | 著者 | Cao, C, Kwon, K, Jiang, Y.L, Drohat, A.C, Stivers, J.T. | | 登録日 | 2003-05-02 | | 公開日 | 2003-11-25 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure and base perturbation studies reveal a novel mode of alkylated base recognition by 3-methyladenine DNA glycosylase I
J.Biol.Chem., 278, 2003
|
|
5FF8
 
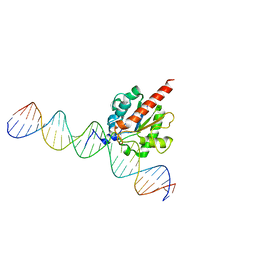 | | TDG enzyme-product complex | | 分子名称: | DNA, G/T mismatch-specific thymine DNA glycosylase | | 著者 | Pozharski, E, Malik, S.S, Drohat, A.C. | | 登録日 | 2015-12-18 | | 公開日 | 2016-09-28 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural basis of damage recognition by thymine DNA glycosylase: Key roles for N-terminal residues.
Nucleic Acids Res., 44, 2016
|
|
5EUG
 
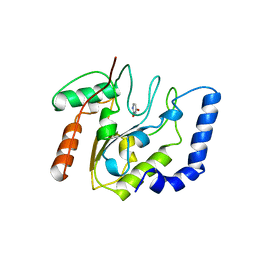 | | CRYSTALLOGRAPHIC AND ENZYMATIC STUDIES OF AN ACTIVE SITE VARIANT H187Q OF ESCHERICHIA COLI URACIL DNA GLYCOSYLASE: CRYSTAL STRUCTURES OF MUTANT H187Q AND ITS URACIL COMPLEX | | 分子名称: | PROTEIN (GLYCOSYLASE), URACIL | | 著者 | Xiao, G, Tordova, M, Drohat, A.C, Jagadeesh, J, Stivers, J.T, Gilliland, G.L. | | 登録日 | 1998-12-27 | | 公開日 | 1999-07-23 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal structure of Escherichia coli uracil DNA glycosylase and its complexes with uracil and glycerol: structure and glycosylase mechanism revisited.
Proteins, 35, 1999
|
|
5HF7
 
 | | TDG enzyme-substrate complex | | 分子名称: | DNA (28-MER), G/T mismatch-specific thymine DNA glycosylase | | 著者 | Pozharski, E, Malik, S.S, Drohat, A.C. | | 登録日 | 2016-01-06 | | 公開日 | 2016-09-28 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | Structural basis of damage recognition by thymine DNA glycosylase: Key roles for N-terminal residues.
Nucleic Acids Res., 44, 2016
|
|
4LND
 
 | | Crystal structure of human apurinic/apyrimidinic endonuclease 1 with essential Mg2+ cofactor | | 分子名称: | DNA-(apurinic or apyrimidinic site) lyase, MAGNESIUM ION | | 著者 | Manvilla, B.A, Pozharski, E, Toth, E.A, Drohat, A.C. | | 登録日 | 2013-07-11 | | 公開日 | 2013-11-27 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Structure of human apurinic/apyrimidinic endonuclease 1 with the essential Mg(2+) cofactor.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3EUG
 
 | | CRYSTAL STRUCTURE OF ESCHERICHIA COLI URACIL DNA GLYCOSYLASE AND ITS COMPLEXES WITH URACIL AND GLYCEROL: STRUCTURE AND GLYCOSYLASE MECHANISM REVISITED | | 分子名称: | GLYCEROL, PROTEIN (GLYCOSYLASE) | | 著者 | Xiao, G, Tordova, M, Jagadeesh, J, Drohat, A.C, Stivers, J.T, Gilliland, G.L. | | 登録日 | 1998-10-13 | | 公開日 | 1999-10-13 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.43 Å) | | 主引用文献 | Crystal structure of Escherichia coli uracil DNA glycosylase and its complexes with uracil and glycerol: structure and glycosylase mechanism revisited.
Proteins, 35, 1999
|
|
5JXY
 
 | | Enzyme-substrate complex of TDG catalytic domain bound to a G/U analog | | 分子名称: | DNA (28-MER), G/T mismatch-specific thymine DNA glycosylase | | 著者 | Pidugu, L.S, Pozharski, E, Malik, S.S, Drohat, A.C. | | 登録日 | 2016-05-13 | | 公開日 | 2016-09-28 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | Structural basis of damage recognition by thymine DNA glycosylase: Key roles for N-terminal residues.
Nucleic Acids Res., 44, 2016
|
|
6U17
 
 | | Human thymine DNA glycosylase bound to DNA with 2'-F-5-carboxyl-dC substrate analog | | 分子名称: | ACETATE ION, DNA (28-MER), DNA (30-MER), ... | | 著者 | Pidugu, L.S, Pozharski, E, Drohat, A.C. | | 登録日 | 2019-08-15 | | 公開日 | 2019-11-20 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Excision of 5-Carboxylcytosine by Thymine DNA Glycosylase.
J.Am.Chem.Soc., 141, 2019
|
|
6U16
 
 | | Human thymine DNA glycosylase N140A mutant bound to DNA with 5-carboxyl-dC substrate | | 分子名称: | 1,2-ETHANEDIOL, DNA (28-MER), G/T mismatch-specific thymine DNA glycosylase | | 著者 | Pidugu, L.S, Pozharski, E, Drohat, A.C. | | 登録日 | 2019-08-15 | | 公開日 | 2019-11-20 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Excision of 5-Carboxylcytosine by Thymine DNA Glycosylase.
J.Am.Chem.Soc., 141, 2019
|
|
6U15
 
 | |
3UFJ
 
 | | Human Thymine DNA Glycosylase Bound to Substrate Analog 2'-fluoro-2'-deoxyuridine | | 分子名称: | 5'-D(*CP*AP*GP*CP*TP*CP*TP*GP*TP*AP*CP*GP*TP*GP*AP*GP*CP*AP*GP*TP*GP*GP*A)-3', 5'-D(*CP*CP*AP*CP*TP*GP*CP*TP*CP*AP*(UF2)P*GP*TP*AP*CP*AP*GP*AP*GP*CP*TP*GP*T)-3', G/T mismatch-specific thymine DNA glycosylase | | 著者 | Pozharski, E, Maiti, A, Drohat, A.C. | | 登録日 | 2011-11-01 | | 公開日 | 2012-04-25 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.967 Å) | | 主引用文献 | Lesion processing by a repair enzyme is severely curtailed by residues needed to prevent aberrant activity on undamaged DNA.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7TC3
 
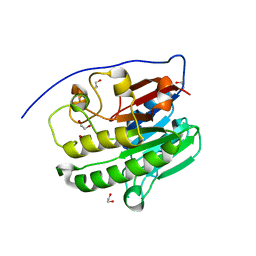 | | Human APE1 in the apo form | | 分子名称: | 1,2-ETHANEDIOL, DNA-(apurinic or apyrimidinic site) endonuclease, mitochondrial | | 著者 | Pidugu, L.S, Pozharski, E, Drohat, A.C. | | 登録日 | 2021-12-22 | | 公開日 | 2022-12-21 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.252 Å) | | 主引用文献 | Characterizing inhibitors of human AP endonuclease 1.
Plos One, 18, 2023
|
|
7TC2
 
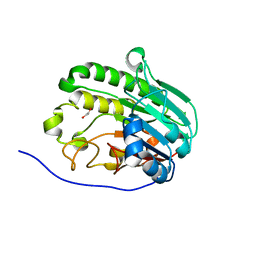 | | Human APE1 in complex with 5-nitroindole-2-carboxylic acid | | 分子名称: | 1,2-ETHANEDIOL, 5-nitro-1H-indole-2-carboxylic acid, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Pidugu, L.S, Pozharski, E, Drohat, A.C. | | 登録日 | 2021-12-22 | | 公開日 | 2022-12-21 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.43 Å) | | 主引用文献 | Characterizing inhibitors of human AP endonuclease 1.
Plos One, 18, 2023
|
|
4XEG
 
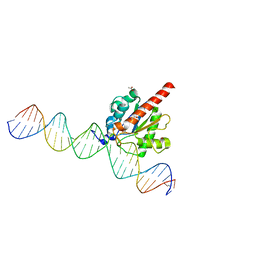 | | Structure of the enzyme-product complex resulting from TDG action on a G/hmU mismatch | | 分子名称: | 1,2-ETHANEDIOL, ACETIC ACID, DNA (28-MER), ... | | 著者 | Pozharski, E, Malik, S.S, Drohat, A.C. | | 登録日 | 2014-12-23 | | 公開日 | 2015-09-09 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | Thymine DNA glycosylase exhibits negligible affinity for nucleobases that it removes from DNA.
Nucleic Acids Res., 43, 2015
|
|
4Z47
 
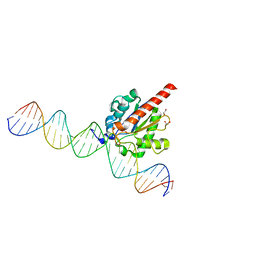 | | Structure of the enzyme-product complex resulting from TDG action on a GU mismatch in the presence of excess base | | 分子名称: | 1,2-ETHANEDIOL, ACETIC ACID, DNA, ... | | 著者 | Pozharski, E, Malik, S.S, Drohat, A.C. | | 登録日 | 2015-04-01 | | 公開日 | 2015-09-16 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Thymine DNA glycosylase exhibits negligible affinity for nucleobases that it removes from DNA.
Nucleic Acids Res., 43, 2015
|
|
4Z3A
 
 | |
